 |

Green Fluorescent Protein
A molecular tag that can be inserted into genes
to make animals and plants glow green.

Timothy King and Paul May
University of Bristol

Molecule of the Month January 2010
Also available: JSMol version.

|
An Introduction to GFP
GFP stands for green fluorescent protein (the official name for the molecule) and is, imaginatively, a protein that fluoresces green in the presence of UV light [1]. It has found its use in all areas of cellular biology and advances have reached the point where it is the focus of works of art [2], such as a pet rabbit called Alba, whose fur glowed green under UV light. In November 2008, three men, Osamu Shimomura, Martin Chalfie and Roger Tsien, were awarded the Nobel Prize [3-7] in chemistry, "for the discovery and development of green fluorescent protein". So what makes this such a versatile and important molecule?
A Brief Explanation of Bioluminescence and Fluorescence
In the first century AD, Pliny the Elder reported that certain jellyfish produced a glowing light [8]. This was the first report of bioluminescence. So what is bioluminescence? Simply put, it is when a biological organism emits light. A common example of this is in the firefly and similar organisms, as seen in figure 1 [9], where the protein luciferin, and the enzyme luciferase [10], combine with oxygen and ATP, a source of energy in living organisms, to produce light. Another example is of deep sea fish which use a similar system to produce light in a lure, attracting prey that they can then eat.

Figure 1. An example of the luciferin/luciferase reaction.
[Reproduced from Ref. 9].
So how does this differ from fluorescence? In fluorescence, incident light is merely re-emitted at a different, less energetic wavelength. Fluorescence works when a photon collides with a molecule, causing the molecule to gain energy. If this energy is enough, an electron will be excited from its initial ground state to a higher energy state, with any excess energy causing the molecule to vibrate more or move faster. The excited molecule slowly loses the vibrational and translational energy, most commonly by colliding with surrounding molecules, or a rearrangement of the structure, but sometimes by other methods. After a length of time, roughly a few microseconds, the molecule can lose the energy stored in the excited electron by allowing it to fall back to its ground state. With this fall, the electron emits a photon with the same energy as the difference between the excited and ground states. Because of the loss of vibrational and translational energy, the energy of absorbed and emitted photons differ, changing the colour of the light emitted [11], e.g. the absorption of blue light and emission of green.
A History of GFP
Osamu Shimomura is the starting point for GFP. As a young boy, he was only miles from the atomic bomb that landed on Nagasaki, close enough to be temporarily blinded by the explosion [3]. In 1960, Osamu Shimomura moved from Japan to work at Princeton University and here he worked in a research group studying the jellyfish Aequorea victoria. The project he was working on involved bioluminescence, the edges of the skirt, or umbrella, of this species of jellyfish emitting a green light, figure 2. So, along with two colleagues from the university he went to Puget Sound, a series of waterways extending into northern Washington State from the Pacific Ocean. The three scientists collected roughly 10,000 specimens of the jellyfish, all with hand nets to avoid catching anything else, from which they removed a 5mm-thick ring from around the skirt, responsible for the glow observed. Somewhat gruesomely, these rings were crushed to remove the liquid constituents and forced through cheese-cloth to separate the wanted material from the solid that made up the jellyfish.
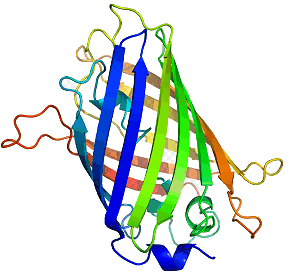
Figure 2. The ring of fluorescence seen in the jellyfish A. victoria.
[Reproduced from Ref. 9].
The crushed rings, or "squeezate" as it was referred to in the original paper, was purified to produce just over 5 mg of glowing substance, though this proved not be the green light observed, but possessed a blue glow instead, originating from a protein named Aequorin. In the purification process another, discarded, protein was found that did posses this green fluorescence, but was practically ignored as it only emitted the distinctive green light when irradiated with blue or UV light. In 1962 the results of the work was published, with only a minor mention of the fluorescing green protein [10].
Realising the potential of the green fluorescing protein, Shimomura shifted his focus, and over the next 17 years continued to work on the jellyfish. In this time, over 850,000 of the organisms were captured and killed. During the 17 years, and due to the increase in numbers, family members were roped in to help, and the scissors that had originally been used to remove the fluorescent skirt were discarded in favour of a specially made jellyfish cutter [3]. At the end of this, Shimomura had finally deciphered the structure of the fluorescing portion of the molecule[12].
Research into GFP slowed considerably until in 1992 the gene for GFP was eventually coded by Douglas Prasher [13], genes and DNA, in general, being the blueprints from which proteins are synthesised. This was a significant breakthrough and led to a race to be the first research teams to successfully express the gene in and organisms. The first person to manage this was Martin Chalfie and in 1994 published the results of this work [14]. Chalfie succeeded in inserting the gene for GFP into the bacteria E. coli, which then fluoresce with a green light in the presence of UV radiation.
Since then, GFP has been used for thousands of different applications, and improvements have been made to the properties of the protein by mutations in the gene, creating brighter variants, as well as multiple different colours. As well as this, GFP and GFP variants have been found in dozens of different marine species, from sea anemones to sea pansies, which produce their own light, to corals that possess no bioluminescence, but have the ability to fluoresce. And the natural varieties do not just stop at green, with almost all imaginable colours having been found [15].
What is the structure of GFP and how does it work?
The structure of GFP is built up in the same way as any protein and as such has multiple levels of structure, as well as multiple methods of chemical interaction. The base or primary structure of GFP is a chain of 238 amino acids [16] weighing roughly 27,000 atomic mass units (27 kDa) [12], with only about 4 of the amino-acids directly producing any fluorescence effect [8]. The secondary structure is a series of helices and pleated sheets, caused by hydrogen-bonding within the chain, while the tertiary structure is a barrel made from 11 of the sheets, capped with the helices. At the centre of this lies the chromophore, a short chain of altered amino-acids responsible for the light emission. The barrel structure keeps the chromophore away from solvents, making GFP capable of fluorescing under almost any conditions, being able to fluoresce nearly to the point at which the protein is denatured by things such as heat and pH [8]. Figure 3 shows the structure of GFP, the cylinder being ~42 Å long and ~24 Å in diameter [17].
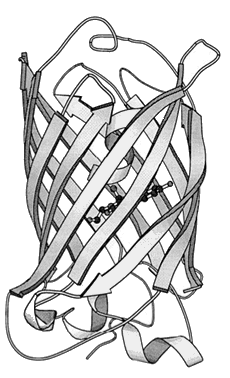
Figure 3. The tertiary structure of GFP. The central darker circles represent the chromophore,
while the long flat sheets represent the barrel surrounding it.
[Reproduced from Ref. 19].
When a protein is produced by a cell, the only part that is synthesised is a chain of amino-acids with no more structure than that. This is referred to as the "primary" structure (1°). Once completed, the amino-acid chain is folded into the right shape, forming the necessary bonds in order to hold the structure rigid. In the case of GFP, this folding brings the amino acids necessary for the chromophore close enough together to enable it to react in a way as to produce the actual chromophore [18]. Scheme 1 shows how the amino-acids form the structure needed for the molecule to fluoresce [19].
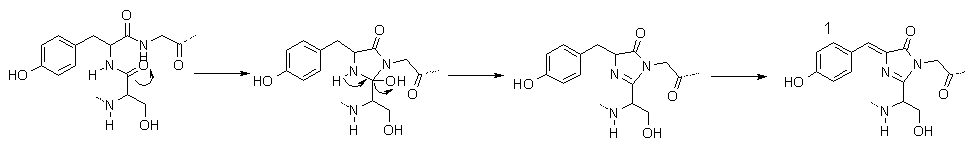
Scheme 1. The mechanism for formation of the chromophore of GFP.
Molecule 1 is the final structure of the chromophore.
The first two steps happen automatically without outside input, however, the final step requires atmospheric oxygen.
The dotted lines represent bonds to the rest of the GFP protein.
So now that we know what GFP looks like, how exactly does it convert blue light to the green light show? This is, surprisingly, the easy part, even though it may look complex at first. Molecule 1, in Scheme 1 shows the complete structure for the part of the protein responsible for fluorescence. To get the jargon out of the way, there are two sets of states that contribute most to the strongest absorptions and emissions, i.e. the UV absorption and green emission. The first state, shown as 1 in scheme 1, is referred to as the A state, or simply A. The other partner in the set is A*, an energetic version of A after absorption of a photon. The second set of states are called I and I*, and have a similar structure to 1, but the phenolic hydrogen (the one on the OH at the left of 1 in Scheme 1), has migrated to an oxygen not shown on the diagram, along a series of hydrogen-bonding links [20, 21], again, with I being the ground state and I* being the energetically excited state.
So now that all of that is out of the way, the way UV light is converted to green is simple. A photon of UV light hits the chromophore, converting the A state to A*. Excess energy from the photon collision converts A* to the slightly lower energy I*, through a simple proton transfer, lowering the energy stored in the system. The I* state then simply releases energy to return to the I state by emitting a photon of green light. The energies of I and A are so close in energy that the proton can simply jump back to its starting place, enabling the whole process to begin again.
Why is GFP important?
So how does all this make GFP a molecule worthy of Noble Prize winning research? Many peptide chains require enzymes to aid in the complex folding that occurs in producing the correct-shaped protein. Many proteins also use enzymes to operate, for example, the luciferin/luciferase combination mentioned earlier. In GFP, however, the complex folding operation occurs automatically and the only thing required for the protein to fluoresce is atmospheric oxygen for the final oxidation of the chromophore, seen in scheme 1.
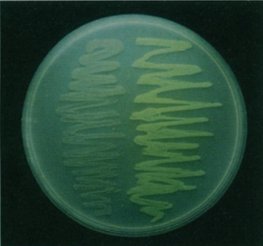
So why is this important? Well this means that organisms, other than those in which GFP naturally occurs, can be genetically engineered to have a gene that produces GFP and it will still work without multiple other genes being implanted [22]. When Chalfie first made glowing green E. coli, it opened wide the possibilities for looking inside a living cell for the first time. Figure 4 shows the Petri dish of glowing E. coli, under a UV light, compared to a regular sample, while figure 5 shows the first example of what makes GFP so amazing. It is a photograph of a round-worm, C. elegans, which has had one of its genes replaced with the genetic code for GFP. This resulted in GFP being expressed in the worm, but only in the places in which the original gene would have been expressed.
 |
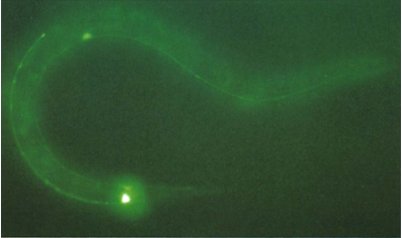 |
Figure 4. A photograph of the genetically altered E. coli (right)
next to a non-altered variety.
[Reproduced from Ref. 14] |
Figure 5. C. elegans with GFP replacing a touch receptor protein.
The bright spots are where the protein would normally be most expressed.
[Reproduced from Ref. 22]. |
Neither organism suffered toxic effects from the protein and the experiment showed that both prokaryotes (bacteria) and eukaryotes (almost everything else) can be made to express GFP. Since then it has been used in organisms as diverse as fruit flies, mice [23], rabbits, tobacco plants and human cells [8]. As well as replacing genes, the relatively small size of GFP, for a protein, enables it to be used as a tag, or reporter gene [24], involving adding the genetic code for GFP onto the end of the gene for the protein that needs to be tagged and growing the organism. This results is the protein being produced with a small tag that doesn't affect the organism or function of the protein at all. The protein can then be seen, identifiable by the green fluorescence enabling the pin pointing of genetic expression.
So far, the examples given have only been concerned with positioning of gene expression, but GFP can be used to do much more. A technique known as FRET [25] (fluorescent resonance energy transfer), can be used to image real-time events within a cell. The basic principle is that two variants of GFP, that absorb and emit light at different wavelengths to each other, are bound to interacting proteins. When these proteins are far apart and the system is illuminated with a light that only excites one of the GFP variants, only the colour from that protein is emitted. However, when the two proteins interact, such as an enzyme acting upon its substrate, and the same light is used, the two GFP variants are brought close enough together for the energy from the absorbed light to be transferred between the GFP molecules, resulting in the colour emitted to change to that of the second GFP.
How has it been improved?
When Roger Tsien first heard about the coding of the GFP gene, instead of wondering how he could implant it into an organism, he wondered how he could make it better - the GFP protein acquired from A. victoria (known as wild-type or wtGFP), sometimes being less than ideal for research. For instance, wtGFP has broad excitation peaks, that is, they absorb multiple colours of light, making it unsuitable for FRET [26]. They are also slow in the formation of the chromophore, taking over 2 hours for the final oxidation to occur. They also have tendency to form dimers and trimers, increasing the molecule in weight massively, which can inhibit not only the function of proteins they are attached to but also the function of the GFP itself. The increased size of the dimerised and trimerised GFP molecules can inhibit the movement of tagged proteins around the cell and through membranes.
So how could you go about solving these? The easiest way to do this, and indeed, the way it was done, is to engineer colonies of bacteria to express the gene and allow them to grow, letting nature produce mutant varieties of GFP in time. In the experiment undertaken by Roger Tsien, a large proportion of these mutations destroyed the fluorescence, but a small fraction resulted in improvements. These included variants that shifted the excitation and emission peaks, changing the colours, variants that replace the two excitation peaks by one, increasing the brightness, and ones that oxidise much faster. As well as GFP mutations, mutations in a similar protein found in a type of coral, Discosoma, called RFP [27] as it fluoresces red, have resulted in a wide spectrum of usable fluorescent proteins, shown in figure 4.
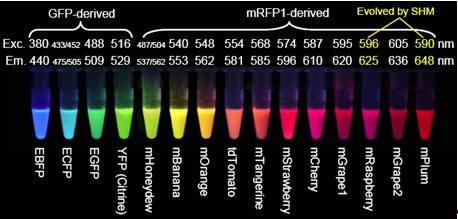
Figure 6. The wide varieties of GFP and variants that have been produced.
mRFP1 is an already altered version of the Discosoma protein.
Exc. stands for excitation and is the peak wavelength at which excitation occurs,
while Em. is the peak emission wavelength.
E before the name of protein signifies enhanced brightness.
[Reproduced from Ref. 26].
Conclusions
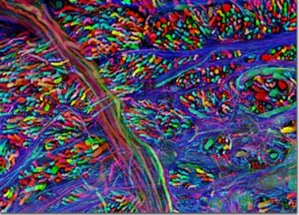
So what is left for GFP and its derivatives? After changing the face of the biosciences and being the subject of a Nobel Prize, GFP has begun to reach the pinnacle of what can be done in terms of new techniques and mutations. However, this does not mean that what has been done will not continue to be done and, with GFP being the powerful tool that it is, continue to change the face of the biosciences. One way in which this is clearly shown is the "brainbow", shown in figures 5 [28] and 6 [29]. Both images are of the nervous systems of animals and both were part of a photography competition, run by Olympus America Inc. The "brainbow"[30] as it is so aptly named, is created by enabling 3 or 4 variants of the GFP and RFP proteins to be produced and act as tags for different proteins used throughout the nerves and brain. Where only one of these proteins is expressed, only that colour is shown, but often, multiple proteins will be expressed in different quantities in the same cells. This combination of GFP molecules produces a rainbow of colours in a similar way to the 3 colours found in printers, allowing the nerve cells in an animal to be seen.
 |
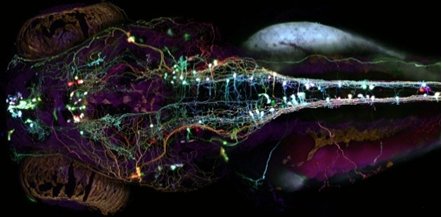 |
Figure 7. Brainbow of a mouse's brain-stem.
[Reproduced from Ref. 29]. |
Figure 8. An image of the Brainbow of a Zebrafish's nervous system.
[Reproduced from Ref. 28]. |
Acknowledgements
Thank you to Dr. Charles King for making sure that the writing was both understandable and concise.

References
- J. W. Hastings and J. G. Morin, Biol. Bull., 1969, 137, 402.
- M. Suvakovic, Politics and art: After the fall of the Berlin wall, Filozofski Vestnik, 2008, 29, 91.
- L. Brindley, A glowing green Nobel, Chemistry World, 2008, 5, 38-40.
- B. G. Levi, Physics Today, 2008, 61, 20-22.
- A. Miyawaki, Cell, 2008, 135, 987-990.
- L. Teodori, T. Bushnell, L. Campanella and A. Tarnok, Cytometry Part A, 2008, 73A, 1109-1110.
- P. S. Weiss, ACS Nano, 2008, 2, 1977-1977.
- A. B. Cubitt, R. Heim, S. R. Adams, A. E. Boyd, L. A. Gross and R. Y. Tsien, Trends in Biochem. Sci., 1995, 20, 448-455.
- O. Shimomura, Nobel Prize website.
- O. Shimomura, F. H. Johnson and Y. Saiga, J. of Cell. and Comp. Physio., 1962, 59, 223.
- A.J. Orr-Ewing, in Photochemistry, University of Bristol, Bristol, 2008.
- O. Shimomura, Febs Letters, 1979, 104, 220-222.
- D. C. Prasher, V. K. Eckenrode, W. W. Ward, F. G. Prendergast and M. J. Cormier, Gene, 1992, 111, 229-233.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802-805.
- G. Mocz, Marine Biotechn., 2007, 9, 305-328.
- R. Hein and R. Y. Tsien, Current Biol., 1996, 6, 178-182.
- M. Ormo, A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien and S. J. Remington, Science, 1996, 273, 1392-1395.
- B. Indge, M. Baker and M. Rowland, A New Introduction to Human Biology (AQA Specification A.), Hodder Murray, 2000.
- R. Y. Tsien, Ann. Rev. Biochem., 1998, 67, 509-544.
- T. Andruniow, International Conference and Workshop on Modeling and Design of Molecular Materials, Wroclaw, Poland, 2006.
- A. Sinicropi, T. Andruniow, N. Ferre, R. Basosi and M. Olivucci, J. Am. Chem. Soc., 2005, 127, 11534-11535.
- M. Chalfie, Current Biol., 1994, 4, 443-443.
- M. Ikawa, K. Kominami, Y. Yoshimura, K. Tanaka, Y. Nishimune and M. Okabe, Development Growth & Differentiation, 1995, 37, 455-459.
- W. Bains, Biotechnology from A to Z, 3rd edn., Oxford University Press, 2004.
- A. Miyawaki, Cell Structure and Function, 2004, 29, 6.
- R. Tsien, http://nobelprize.org/nobel_prizes/chemistry/laureates/2008/tsien-slides.pdf.
- R. E. Campbell, O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias and R. Y. Tsien, Proc. Nati. Acad. Sci. USA, 2002, 99, 7877-7882.
- A. Pan, Larval "Brainbow" Zebrafish, http://www.olympusbioscapes.com/gallery/2008/4.html.
- J. Livet, "Brainbow" Mouse Brain Stem, http://www.olympusbioscapes.com/gallery/2007/index.html.
- J. Livet, T. A. Weissman, H. N. Kang, R. W. Draft, J. Lu, R. A. Bennis, J. R. Sanes and J. W. Lichtman, Nature, 2007, 450, 56.


 Back to Molecule of the Month page. [DOI:10.6084/m9.figshare.5255080]
Back to Molecule of the Month page. [DOI:10.6084/m9.figshare.5255080]

![]()
![]()
![]()
![]()








