Matrix-assisted Laser Desorption/Ionisation (MALDI)
In the early 1960's, it was demonstrated that the irradiation of
low-mass organic molecules with a high-intensity laser pulse formed
ions that could be successfully mass analysed. This was the origins of
laser desorption (LD) ionisation. Other the next few decades, the
technique underwent substantial development, culminating in the
extension of the technique to the volatilisation of non-volatile
biopolymers and organic macromolecules. There was, however, a sharp
cut-off in mass at about 5-10 kDa, limiting the technique's
application. The other main limitation was that ions were created in
bursts which prevented the technique from being coupled to scanning
mass analysers. In fact LD was only really successful when coupled to
time-of-flight (TOF) mass analysers.
In 1987, Michael Karas and Franz Hillenkamp [1] successfully demonstrated the use of a matrix (a small organic molecule) in LD to circumvent the mass limitation. The matrix had a strong absorbance at the laser wavelength and was highly sublimable [2]. A low concentration of the analyte was mixed with this matrix onto a probe or metal plate (see figure) and introduced into a pulsed laser beam. A substantial burst of ions was produced with each laser pulse. An unexpected side effect of the matrix was that it allowed for the laser incidence spot to be refreshed between each pulse, thus greatly enhancing shot-to-shot reproducibility. This was the foundation of matrix-assisted laser desorption/ionisation (MALDI). Later developments by Koichi Tanaka [3] demonstrated the application of MALDI to a whole range of biological macromolecules. This gave him a part share of the 2002 Noble prize for chemistry [4], making him the 5th mass spectrometry pioneer to receive such an honour.
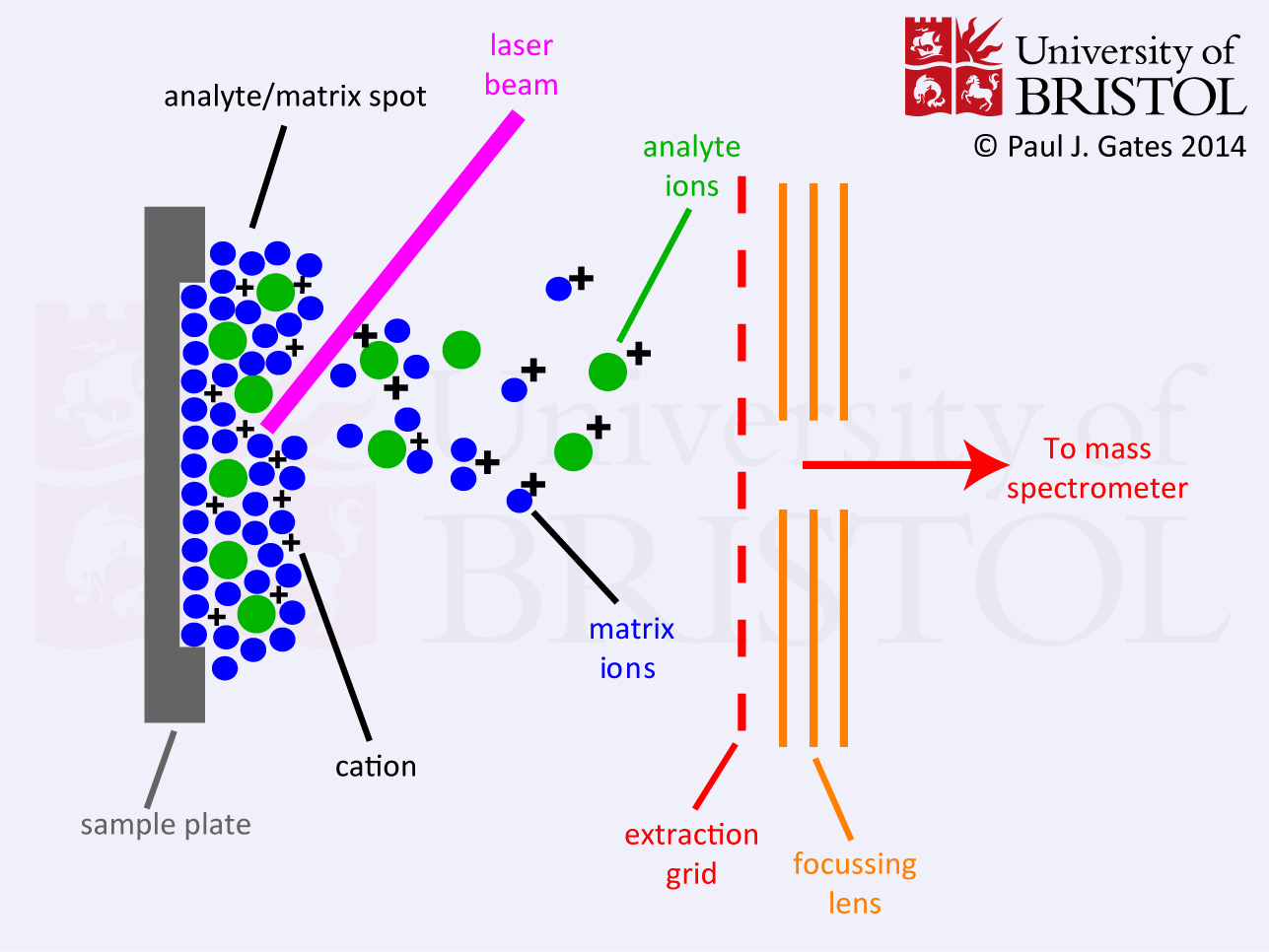
The mechanism of MALDI is believed
to consist of three basic steps:
(i) Formation of a 'Solid Solution': It is essential for the matrix to be in excess thus leading to the analyte molecules being completely isolated from each other. This eases the formation of the homogenous 'solid solution' required to produce a stable desorption of the analyte.
(ii) Matrix Excitation: The laser beam is focused onto the surface of the matrix-analyte solid solution. The matrix chromophore absorbs the lase irradiation causing rapid vibrational excitation, bringing about localised disintegration of the solid solution. The clusters ejected from the surface consist of analyte molecules surrounded by matrix and salt ions. The matrix molecules evaporate away from the clusters to leave the free analyte in the gas-phase.
(iii) Analyte Ionisation: The photo-excited matrix molecules are stabilised through proton transfer to the analyte. Cation attachment to the analyte is also encouraged during this process. It is in this way that the characteristic [M+X]+ (X= H, Na, K etc.) analyte ions are formed. These ionisation reactions take place in the desorbed matrix-analyte cloud just above the surface.
[2] A series of papers covering matrix development were published shortly after the initial MALDI papers - for example:
R.C. Beavis and B.T. Chait, Rapid Communications in Mass Spectrometry, 3, 1989, p432.
R.C. Beavis and B.T. Chait, Organic Mass Spectrometry, 27, 1992, p156.
M. Karas and F. Hillenkamp, Organic Mass Spectrometry, 28, 1993, p1476.
[4] K. Tanaka, Angewandte Chemie - International Edition, 42, 2003, p3861.
[5] Several good reviews of MALDI have also been published - for example:
M. Karas et. al., Mass Spectrometry Reviews, 10, 1991, p335.
R.C. Beavis, Organic Mass Spectrometry, 27, 1992, p653.
U. Bahr, M. Karas and F. Hillenkamp, Fresenius' Journal of Analytical Chemistry, 348, 1994, p783.
In 1987, Michael Karas and Franz Hillenkamp [1] successfully demonstrated the use of a matrix (a small organic molecule) in LD to circumvent the mass limitation. The matrix had a strong absorbance at the laser wavelength and was highly sublimable [2]. A low concentration of the analyte was mixed with this matrix onto a probe or metal plate (see figure) and introduced into a pulsed laser beam. A substantial burst of ions was produced with each laser pulse. An unexpected side effect of the matrix was that it allowed for the laser incidence spot to be refreshed between each pulse, thus greatly enhancing shot-to-shot reproducibility. This was the foundation of matrix-assisted laser desorption/ionisation (MALDI). Later developments by Koichi Tanaka [3] demonstrated the application of MALDI to a whole range of biological macromolecules. This gave him a part share of the 2002 Noble prize for chemistry [4], making him the 5th mass spectrometry pioneer to receive such an honour.

(i) Formation of a 'Solid Solution': It is essential for the matrix to be in excess thus leading to the analyte molecules being completely isolated from each other. This eases the formation of the homogenous 'solid solution' required to produce a stable desorption of the analyte.
(ii) Matrix Excitation: The laser beam is focused onto the surface of the matrix-analyte solid solution. The matrix chromophore absorbs the lase irradiation causing rapid vibrational excitation, bringing about localised disintegration of the solid solution. The clusters ejected from the surface consist of analyte molecules surrounded by matrix and salt ions. The matrix molecules evaporate away from the clusters to leave the free analyte in the gas-phase.
(iii) Analyte Ionisation: The photo-excited matrix molecules are stabilised through proton transfer to the analyte. Cation attachment to the analyte is also encouraged during this process. It is in this way that the characteristic [M+X]+ (X= H, Na, K etc.) analyte ions are formed. These ionisation reactions take place in the desorbed matrix-analyte cloud just above the surface.
References:
[1] M. Karas and F. Hillenkamp, International Journal of Mass Spectrometry and Ion Processes, 78, 1987, p53.[2] A series of papers covering matrix development were published shortly after the initial MALDI papers - for example:
R.C. Beavis and B.T. Chait, Rapid Communications in Mass Spectrometry, 3, 1989, p432.
R.C. Beavis and B.T. Chait, Organic Mass Spectrometry, 27, 1992, p156.
M. Karas and F. Hillenkamp, Organic Mass Spectrometry, 28, 1993, p1476.
[4] K. Tanaka, Angewandte Chemie - International Edition, 42, 2003, p3861.
[5] Several good reviews of MALDI have also been published - for example:
M. Karas et. al., Mass Spectrometry Reviews, 10, 1991, p335.
R.C. Beavis, Organic Mass Spectrometry, 27, 1992, p653.
U. Bahr, M. Karas and F. Hillenkamp, Fresenius' Journal of Analytical Chemistry, 348, 1994, p783.