Enzymes are large biological molecules which catalyse nearly all chemical reactions in biological systems, by stabilising the transition states. They are also highly specific in both the reaction catalysed and in their choice of reactants27,18.
They are nearly all proteins, which are built up from amino acid residues. Amino acids are the basic structural units of proteins, and all contain an amino group, a carboxyl group, a hydrogen atom, and a distinguishing R-group (side chain), which are all bonded to an a -carbon
3.2.1 Nomenclature – Designation of the atoms within residues
The central carbon atom on the backbone (or main chain) is called the a -carbon. The other parts of the backbone are known as the a -carbonyl and a -amino groups. The naming on the side chain is a little more complicated. The atoms next to the a -carbon are designated, using the Greek alphabet, on how far away they are from the a -carbon.
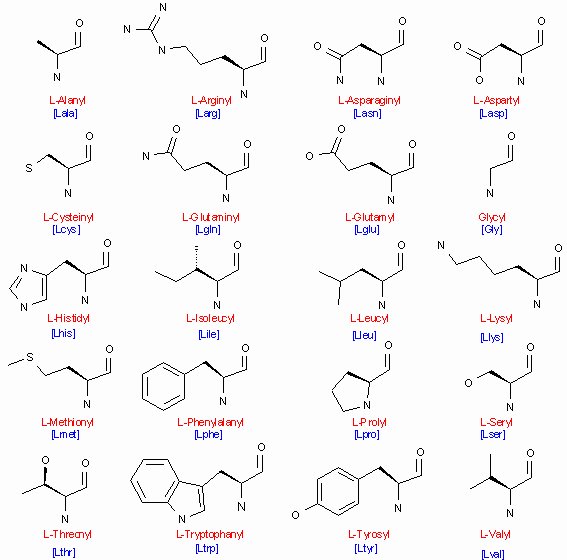
There are 20 different side chains commonly found in proteins (figure 3.2.1a).
Figure 3.2.1a - Formula of 20 the L-amino acids

Proteins are formed from peptide linkages (a -carboxyl to a -amino groups) between adjacent amino acid residues (see figure 3.2.1b). The regularly repeating part is called the mainchain or backbone, while the R groups are known as the side chains. Most enzymes contain over 100 amino acid residues. Other types of linkages (interactions) also exist between residues and they are disulphide bonds, hydrogen bonding, van der Waals, and hydrophobic interactions. These interactions all help to stabilise the enzyme27,18. The most important enzyme activity occurs around the active site. This is a region within the enzyme, usually only a small part, which binds to the substrate. It contains the residues that directly participate in bond formation and breaking, called the catalytic residues. The active site is a 3-D entity and so residues far apart in the linear sequence can be important. The substrates are bound to the ‘cleft’ of the active site by multiple weak bonds. The currently accepted theory on enzyme-substrate binding is the induced fit model of D.E. Koshland Jr5. This model states that the active site assumes a shape that is complementary to the substrate only after it has been bound.
Inhibition of enzymes occurs when a molecule reduces or eliminates the activity of the enzyme. Such a molecule is known as an inhibitor, and can bind either at the active site or another part of the enzyme. Inhibition can be reversible or irreversible27,18.
In general, due to the complexity of enzymes, knowledge of the dynamic behaviour of enzymes, especially the active site, will provide an insight into its structure and function. And aid in the design of inhibitors to the enzymes.