The Manganese-calcium oxide cluster of Photosystem II
and its assimilation by the Cyanobacteria
James D. Johnson M.S.
Key: Hyperlink, Pop-up window Introduction:
This month's MOTM is an inorganic cubane complex known as the manganese-calcium oxide cluster, commonly referred
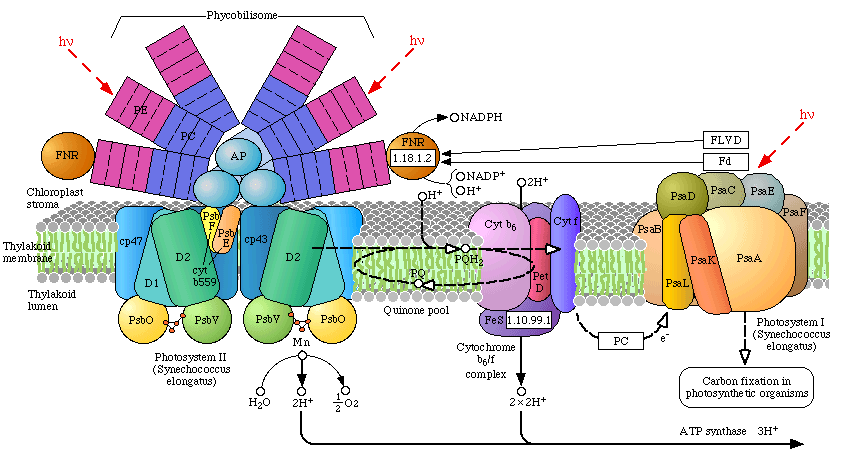
There are two photosystems, PSII and PSI, which communicate with one another
via an electron transport chain known as the "Z scheme" of
photosynthesis. Electrons are initially extracted from water, one at a time (1 photo per electron) at the site of the OEC, and finally released to a soluble ferredoxin
complex after passing through PSI (where it is further re-energized by a second photon). Ferredoxin then reduces Fd-NADP
oxidoreductase, which reduces oxidized NADP, which then reduces 1,3-bisphosphoglycerate. This reduction step regenerates
Ribulose-5-phosphate followed by incorporation of CO2 by Ribulose-bisphosphate carboxylase (Rubisco) -
the most abundant protein on the planet). The reduction of CO2(carbon fixation) is known as the "dark reaction" or Calvin Cycle (sugar production). In algae and plants the thylakoid membranes are found partially stacked (grana) where PSII occurs in higher concentration than PSI (the stacking of the thylakoids may be related to an increase in the photon transfer efficiency by the
surrounding light harvesting chlorophyll complexes - in the pop up of the light harvesting complexes,
notice the concentric arrangement of protein, chlorophyll (and carotenoids), centered around the PSII reaction center. Recently the three dimensional atomic structure of the
manganese-calcium oxide complex was determined by X-Ray diffraction. (3)The
manganese cluster is essentially an inorganic cubane (cubical) structure
found in inorganic crystals made of ranciéite
or hollandite
[Mn4CaO9*3H2O].
These minerals are believed to have been assimilated during the early
Archaean period by the Cyanobacteria approximately 3,200 - 2,800 million
years ago (or 3.2 - 2.8 Ga=giga years ago). The assimilation of
the manganese-calcium oxide complex by the Cyanobacteria is one of
several examples of metallo-catalysts incorporated by living systems.
Two other inorganic metallo complexes assimilated by early prokaryotes
(and are now ubiquitous redox centers in living systems) include the
cubic iron-sulfur centers [Fe4-S4]
[image]
(originating as greigite
(Fe
Alumnus, Department of Chemistry
Florida State University, Tallahassee, FL, USA
June 2006
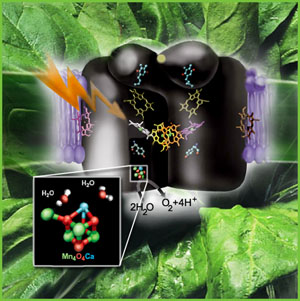 to as the "Oxygen Evolving Complex" or OEC (also referred to as a water oxidase). The OEC is found on the oxidizing side of
Photosystem II (PSII), and is located within organelles known as chloroplasts
in all plants and algae. The OEC is also found in one group of bacteria, the Cyanobacteria (formally called the "blue-green algae", a
name now discounted since the Cyanobacteria are now known not to be prokaryotic bacteria). It is believed that the Cyanobacteria are the
endosymbiotic ancestors of modern day chloroplasts. (1)
In the case of higher plants and algae, the OEC is found integrally embedded within the thylakoid membranes,
located throughout a plant cell's chloroplasts. Cyanobacteria have no internal organelles, and therefore no chloroplasts, and the OEC is found
within thylakoid membranes distributed throughout the cytoplasm.
The Cyanobacteria are quite small, and are about the same size as the internal chloroplasts of the eukaryotic plants and algae. This is one
branch of evidence which supports the theory that chloroplasts are ancient Cyanobacteria (many other facts exists which also support this
theory (circular DNA, bacterial ribosomes, et al.). To review the components of PSII, click on the image at left to explode the details of
the PSII reaction center. The image above was obtained from a review article on PSII published in Science Beat,
an on-line science newsletter produced by Berkeley University (2).
to as the "Oxygen Evolving Complex" or OEC (also referred to as a water oxidase). The OEC is found on the oxidizing side of
Photosystem II (PSII), and is located within organelles known as chloroplasts
in all plants and algae. The OEC is also found in one group of bacteria, the Cyanobacteria (formally called the "blue-green algae", a
name now discounted since the Cyanobacteria are now known not to be prokaryotic bacteria). It is believed that the Cyanobacteria are the
endosymbiotic ancestors of modern day chloroplasts. (1)
In the case of higher plants and algae, the OEC is found integrally embedded within the thylakoid membranes,
located throughout a plant cell's chloroplasts. Cyanobacteria have no internal organelles, and therefore no chloroplasts, and the OEC is found
within thylakoid membranes distributed throughout the cytoplasm.
The Cyanobacteria are quite small, and are about the same size as the internal chloroplasts of the eukaryotic plants and algae. This is one
branch of evidence which supports the theory that chloroplasts are ancient Cyanobacteria (many other facts exists which also support this
theory (circular DNA, bacterial ribosomes, et al.). To review the components of PSII, click on the image at left to explode the details of
the PSII reaction center. The image above was obtained from a review article on PSII published in Science Beat,
an on-line science newsletter produced by Berkeley University (2).
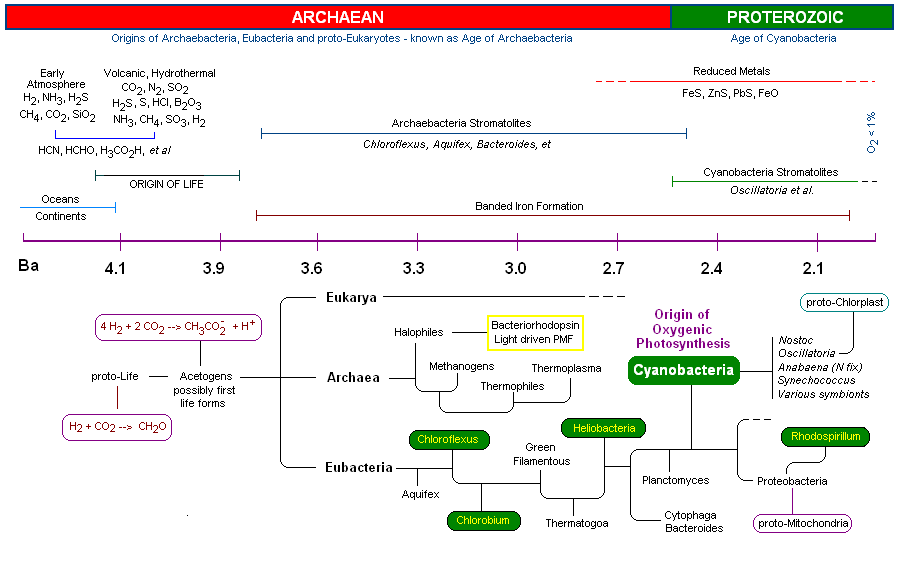
The successful assimilation of the OEC by early Cyanobacteria resulted in an explosion of this group which became the predominant life form for 2 billion years. Unlike other photosynthetic bacteria (discussed briefly below) the Cyanobacteria are "oxygenic photosynthesizers" capable of oxidizing water as a substitute for other electron donors such as H2S and reduced metals (Fe(II) e.g.), producing molecular oxygen as a by-product. Oxygen irreversibly changed the course of life on earth by releasing O2 in great quantities (changing, for example, the composition of the atmosphere as well as earth's geology). During this time the Cyanobacteria successfully dispersed globally and diversified through evolutionary speciation. (5) Finally, member(s) of the Cyanobacteria symbiotically merged with eukaryotes at about 2.0 Ga to become the chloroplast of all algae and higher plant groups on the earth today. The arrival of the OEC of PSII resulted in the most profound impact on the history of evolution. Indeed, the OEC is single-handedly responsible the rise of all plants and animals on the earth today.
1.
First Life - the extremophilic Archaebacteria (4.0 - 3.5 Ga)
2.
Bacterial Photosynthesis (3.5 - 2.7 Ga)
3.
The Oxygen Evolving Complex (OEC) is assimilated by Cyanobacteria (2.7 - 2.5 Ga)
4. The molecular structure of the manganese-calcium oxide water splitting complex
1. First Life - The Extremophilic Archaebacteria (4.0 - 3.5 Ga)
Early Earth:
It is estimated that earth's oceans, continents and atmosphere required approximately 600 million years to stabilize (4.6 - 4.0 Ga) to
the point where liquid water accumulation and continental crust accretion were possible. (3)
Some reports put this time as early as 4.4 Ga. (3).
Early chaotic surface events, though becoming less frequent with time, included meteor bombardment, unstable temperatures, strong lunar forces, tides and storms, risings oceans, reduced sunlight intensity, short day length, UV,
X-Ray and other high energy irradiation, volcanic and geologic upheavals and an ever changing atmospheric chemistry (6). Earth's atmosphere at the beginning of the Hadean period
(4.6 - 4.2 Ga) was strictly reducing (lacked oxygen) (7),
having measurable levels of H Fig 1. is an illustration of the earth relatively soon after it coalesced. Surface temperatures would have been very high, as well as meteorite impact frequency (although in time the meteor impact
frequency would decrease to about one impact per century by 4.0 Ga). (5) (Fig 2). This early stage
of the earth's history is known as the Hadean Eon, and it was towards the end of this eon that the continents were formed and
the oceans accumulated (approximately 4.0 Ga).
Fig 1. is an illustration of the earth relatively soon after it coalesced. Surface temperatures would have been very high, as well as meteorite impact frequency (although in time the meteor impact
frequency would decrease to about one impact per century by 4.0 Ga). (5) (Fig 2). This early stage
of the earth's history is known as the Hadean Eon, and it was towards the end of this eon that the continents were formed and
the oceans accumulated (approximately 4.0 Ga).
First Life:
The first organisms were extremophilic chemotropic
prokaryotes (that is, tolerated extreme environmental conditions such as
high heat, high salt concentration, high and low pH, etc). The
extremophiles are now known to have been early members of the domain
Archaebacteria, although it is argued that simultaneous evolution of the
eubacteria as well as the eukaryotes paralleled these early
extremophiles (15). Many species of the
archaebacteria survive
 today (such as the acetogens,
methanogens,
thermophiles
and
halophiles).
These organisms, like true bacteria, lack a nucleus, as well as
organized internal membrane structures, vacuoles and internal
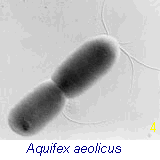
organelles. The bacteria shown in Fig 4 (right) is genetically
positioned as the oldest living fossil on the earth today. [see Time
Chart below]
today (such as the acetogens,
methanogens,
thermophiles
and
halophiles).
These organisms, like true bacteria, lack a nucleus, as well as
organized internal membrane structures, vacuoles and internal
organelles. The bacteria shown in Fig 4 (right) is genetically
positioned as the oldest living fossil on the earth today. [see Time
Chart below]
The early archaebacteria did however possess an amazing array of biochemical pathways to capture energy, acquire carbon, nitrogen, phosphorous, sulfur, hydrogen, bicarbonate and various organics as well as the requisite minerals needed for growth. In addition they had RNA and DNA, giving them the ability to store and pass on genetic information necessary to build enzymes to carry out the multitude of necessary biochemical processes for life (one imagines during this time of early proto-life that modularity, symmetry and gene duplication and transfer were the order of the day (see bacterial conjugation)).
Fundamentally, the early prokaryotes had the capacity (as extant members of the Archaebacteria and Eubacteria have today) to "detect" and "respond" to changes in their environment, such as redox potentials, nutrient levels, physical events, light and temperature variances, et al. (proto-7TM proteins e.g. see Bacteriorhodopsin). The ability to respond biochemically to changes in their immediate environments allowed these organisms to adjust, tolerate, thrive, reproduce and evolve. The various hypothetical pathways which led to earth's first life forms from proto-life is an area of active and intense research. It is believed that the first organisms were acetogens or closely related methanogens [see Timeline below].
The oldest fossils of life on earth are found in microfossils (3.8 Ga) and stromatolites (3.5 Ga). As shown in (Fig 5, 6 and 7) Stromatolites appeared in the early shallow waters of the earth approximately 3.8 Ga. The artist's rendition in Fig. 5 (below, left) of the earth at that time accurately depicts the volcanic outgassing and undersea hydrothermal vent activity which would have been prevalent near the end of the Hadean eon. The earliest stromatolites were probably pre-Photosynthetic eubacteria ancestors of Chloroflex and Chlorobium, both photosynthetic archaean anaerobic thermophiles which preceded the Cyanobacteria.

Although early sedimentary rocks have undergone extensive metamorphosis (thereby destroying microfossil remains), scientists have introduced other techniques to determine where an organism sits on the phylogenetic "tree of life". Biological macromolecules are studied for both function and sequence overlap (genetic homology, divergence, et al.) which has proved especially valuable and has become central to establishing early relationships. Extant biochemical pathways also lend evidence to early relationships, and geochemical biomarkers, molecules and minerals left behind by species specific biological activity (or just biological v. geologic origin), has been used (the recent discovery of sterane, e.g., in 2.7 Ga sediments demonstrated that Eukaryotes were present and that free oxygen was available for biosynthesis). Radioactive analysis may also prove useful for the determination of biochemical activity.
Geochemists who study early life suggest that life may have originated in and around undersea hydrothermal vents (see M. Russell's First Life, referenced below). Hydrothermal sites are excellent locations ideally suited for first life for a number of reasons. Undersea thermal vents provide protection from UV radiation, which would have restricted life form from establishing itself on the surface (whether marine, aquatic or terrestrial). Hydrothermal vents would also have provided long-term stability with regards to a number of parameters, such as pH, temperature and minerals.
As pointed out by Dr. Russell and Dr. Hall (16), hydrothermal vents provide a warm, constant temperature springs (hydrothermal vents have recently been discovered with temperatures near 90 deg C rather than the typical "black smoker" temperature of 400 deg C. - link is to video of the "Lost City" thermal vents in the mid Atlantic). Thermal stability maintained over long periods of time (millions of years) would ideally accommodate the environmental needs of early life . The early Hadean oceans are believed to have been acidic and the effluent emanating from thermal vents are known to be alkaline (perhaps the first open door to a protomotive force, maintaining a constant gradient which early life may have taken advantage of). In the Jan 2006 edition of the journal American Scientist, Dr. Michael Russell presents several models founded on this latter concept.
One of the most attractive ideas of thermal vents as candidates for
nursing first life is the geochemistry of submarine hydrothermal vents. Early in
the history of the earth reduced minerals such as Fe(II) and Ni(II). As these reduced metals entered into
 t
the acidic environment of the sea, the change is temperature may have facilitated precipitation as
carbonates, silicas, clays and iron-nickel sulfides. These precipitates, like stalagmites in a limestone cave, would accumulate and build up around the vents.
It is known that these structures are porous, thereby providing ideal breeding grounds for first life.
t
the acidic environment of the sea, the change is temperature may have facilitated precipitation as
carbonates, silicas, clays and iron-nickel sulfides. These precipitates, like stalagmites in a limestone cave, would accumulate and build up around the vents.
It is known that these structures are porous, thereby providing ideal breeding grounds for first life.
One could imagine as well that perhaps many of these vents were in fairly shallow water and further oxidation and precipitation may have been facilitated by interaction of the metals with UV at the surface. Hydrothermal vents may have been life's first "flow reactor" (as characterized by Dr. Russell); providing a long term stable environment, with plenty of nutrients and minerals, as well as thermal and chemical potential that early proto-life could/may have taken advantage of.
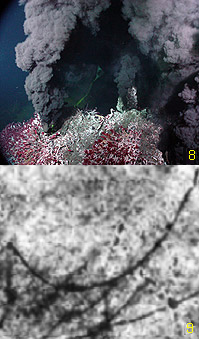
Fig 8, left, shows a black smoker hydrothermal vent. This particular vent may be one of the higher temperature vents (400+ deg C), but perhaps cooler vents, in the vicinity of 100 deg C, may have provided opportunity for early thermophilic prokaryotes to establish themselves. Until recently no fossilized evidence of early prokaryotes had been reported around hydrothermal vents (at least prior to 544 Mya). Fig 9, left, shows an image of a proposed early prokaryote (pyritic filaments) found in a 3.235 Ga volcanic sulphide deposit from the Pilbara Craton of Australia. (x) Dr. Birger Rasmussen, the scientist from the University of Western Australia who reported this find, proposed that this was evidence of a thermophilic chemotropic prokaryote, embedded within a matrix of pyrite (more than likely an iron-oxidizing prokaryote), and inhabited sub-sea-floor hydrothermal environment. This finding would be in line with the proposal that life originated at the base of ancient hydrothermal vents. Though this finding is not absolute proof standing alone (some controversial results were seen in the literature regarding this find) it does represent the efforts that are underway to continue looking for early signs of early life around ancient fossilized venting systems.
By 2.5 Ga, oxygenic photosynthesis was beginning to make a clear mark in geologic sediments. Cyanobacteria, by assimilating the OEC and releasing molecular oxygen, changed the geological landscape of the earth, as well as its atmosphere and oceans. All other organism would respond in abeyance, either taking up oxygen tolerant metabolism or recess into niches devoid of oxygen. The assimilation of the manganese-calcium complex by the cyanobacteria laid the ground work for the Cambrian explosion, and for the evolution of the animal and plant kingdoms.
Previous concepts of taxonomy, based primarily on morphology and physiology, have been supplanted by the work of Woese and others who revolutionized the technique of using DNA as molecular fossils (16). Woese and his group are responsible for the discovery of the domain Archaea, and in the process laid the ground work for future research in this area (Woese's work lends much credence to the old saying "..when one door is closed, many more are open.").
2. Bacterial Photosynthesis (3.8 - 2.7 Ga)
Introduction to the Photosynthetic bacteria: Thirty-six bacterial lineages (Eubacteria) have been identified, of which only five are capable of using chlorophyll-based energy conversion to create a Protonmotive Force (PMF) to drive ATP synthesis and reduce CO2 to sugars. Of the five bacterial groups capable of photoautotrophic photosynthetic growth, four, (the exception being the Cyanobacteria), perform photosynthesis under anaerobic conditions and do not oxidized water to molecular oxygen via the OEC (anoxygenic photosynthesis). Indeed, the Cyanobacteria are the only group of bacteria which have assimilated the OEC necessary for splitting water. Members of the five photosynthetic bacterial families are shown below (these are example species from each group):
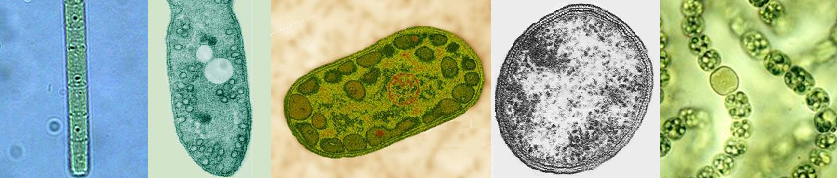
|
| From l to r: Chloroflexus (green non-sulfur bacteria, Chloroflexaceae), Rhodospirillum (purple bacteria, Rhodospirillaceae), Chlorobium (green sulfur bacteria, Chlorobiaceae), Heliobacterium (Gram-positive, Heliobacteriaceae), and Nostoc (Cyanobacteria, Nostocaceae). Note the N2 fixing heterocyst in Nostoc. |
One of the most fascinating areas of biochemistry today is the determination of evolutionary relationships among early prokaryotes (including all three domains, see time line below). Recent findings have revealed that early prokaryotes are highly heterophyletic. Scientists now believe that the transfer of genetic information between divergent organisms has been prolific, especially very early in evolution, thereby complicating further the genomic mapping of their ancestry (genes may be transferred between divergent organisms though such processes such as conjugation, as previously mentioned. These early transformational events have literally turned the "tree of life" into a three dimensional "web of life." With species dying out in the interim, and evolution spanning more then 3,000 million years, bio- and geo-chemical researchers have plenty to keep them busy for the foreseeable future. A time link was compiled while reading the various articles to write this MOTM page (Best estimates are plotted. The diagram has many hyperlinks embedded within it, e.g., you may be interested to read about "The Banded Iron Formation". For the longest time it was believed that these iron oxide patterns in fossilized rock was the result of cyclical release of oxygen by Cyanobacteria. Alternative routes of iron oxidation have been proposed to account for its early appearance - see link).
An excellent exercise to carry out in a biology class is to set up what is called a "Winogradsky Column". The column is designed to demonstrate the interdependence of various bacteria on metabolites and metabolic wastes. Using pond material a large graduated cylinder is filled and allowed to settle for 1-2 months. Within a few weeks three distinct layers begin to separate out, q.v., (1) aerobic water, (2) anaerobic water, and (3) anaerobic sediment. Anaerobic organisms occupy the oxygen free zones and the Cyanobacteria the aerobic zones. The procedure for this exercise can be found at Dr. Jim Deacon's Molecular Biology demonstration site at the University of Edinburgh.
Bacterial reaction center and light harvesting chlorophylls:
All known microorganisms use two functionally unrelated processes for the conversion of light into chemical energy. Chlorophyll-based systems are used by the photosynthetic bacteria to drive photosynthesis, and bacteriorhodopsin (bR), considered a divergent adaptation by the Halophilic prokaryotes (keep in mind that the 7TM sensory peptides appear to be a derivation of this scheme). The Halophiles use light sensitive bacteriorhodopsin to pump protons outside of the cell in order to maintain salt balance in hypersaline environments. Note that the photosynthetic schemes (electron transport) is fundamentally different between the Cyanobacteria, the Purple bacteria and the Green Sulfur bacteria (pop are added for a quick overview but non-oxygenic photosynthesis is not discussed here).
Of the photosynthetic bacteria, three schemes are used for light harvesting pigments. Chlorobium spp. uses a specialized internal membrane bound body known as a Chlorosome (see below). Chloroflexus spp., Heliobacteria spp., and Rhodospirillum spp. use a class of Light Harvesting proteins (below right) in which bacteriochlorophyll is embedded and arranged to focus and transfer light to the bacteria reaction center (below left). Finally, the only bacteria to use the OEC in oxygenic photosynthesis utilizes Phycobilisomes (see below).
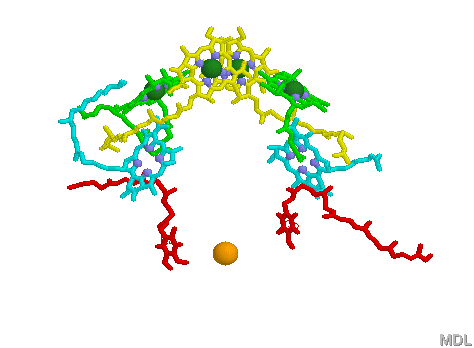
Light Harvesting Chlorophyll: A typical bacterial reaction center is shown below, left. Note the bacteriochlorophyll reaction center "dimer", a special pair of chlorophyll molecules (yellow with green Mg++) designed to absorb incoming photons from surrounding light harvesting chlorophyll complexes. The light blue Mg++ free bacteriochlorophylls shown are pheophytins which act to transfer an electron from the reaction center dimer to a pair of quinones (shown in red). The solitary orange atom represent iron. (see pop up images 2 paragraphs above for more detail).
Surrounding the bacterial reaction center in several bacteria species, algae and plants are complexes known as Light Harvesting proteins (below right). These proteins bind chlorophyll (in this case bacteriochlorophyll) molecules and other pigments such as carotenoids (note that one role carotenoids play is to prevent the reaction center from becoming photo damaged by over oxidation). What is really amazing about the Light Harvesting complexes is the light transferring efficiency from initial absorption until the photon reaches the reaction center special chlorophyll dimer (see efficiency pop-up for a visual demonstration). The chlorophyll molecules which are held within the protein complex (green molecules in image below right) are positioned to maximize photon transfer. The electron path after reaching the dimer moves through the pheophytins (light blue) and on to the quinones (red, below image left). The reaction center dimer is then reduced by an external electron sources such as Cytochrome or, in the case of the Cyanobacteria, algae and plants, the oxygen evolving complex (OEC). A brief review of the various light harvesting arrangements is given below. The PSII complex is much more sophisticated then the simple bacterial light harvesting complex shown below.
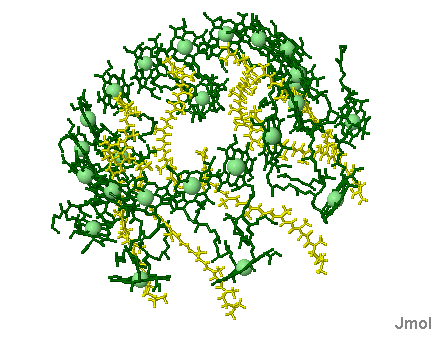
Phycobilisomes: Phycobilisomes are light harvesting pigments found throughout the Cyanobacteria (and in red algae). All other photosynthetic bacteria use the light harvesting complex as described above. Phycobilisomes, which specialize in absorbing deep penetrating green light (more than 1 meter deep, e.g., in marine environments), are capable of re-emitting light in regions which the other photosynthetic pigments can absorb. This would be a very advantageous adaptation in marine environments in which protection from UV radiation might be needed. There is a central core of allophycocyanin (AP) which sits above the photosynthetic reaction center. There are phycocyanin and phycoerythrin subunits which radiate out from this center like thin tubes. The fluorescent pigments which are present in the phycobilisome, such as phycocyanobilin (PC) and phycoerythrobilin (PE) re-emit the green light in regions.
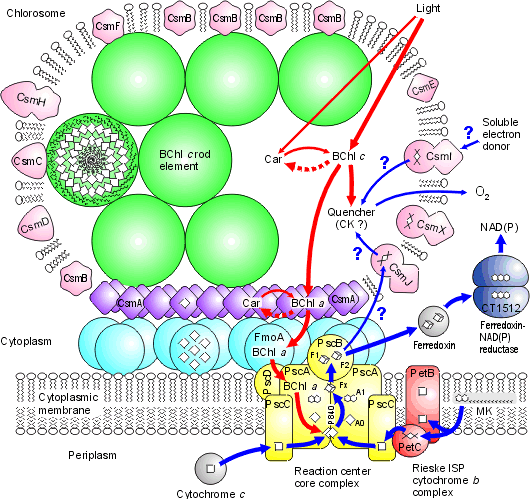
Chlorosomes: Model of the photosynthetic apparatus in Chlorobium tepidum showing "chlorosome" body compiled by Dr. Donald Bryant and associates at Penn State University (Chlorosomes found in the green sulfur bacteria, family Chlorobiaceae). Light and excitation energy transfer is shown in red, electron transfer is shown in blue. Proteins and chlorosome rod elements are shown in color and some cofactors are indicated as white symbols. Diamonds represent individual BChl c molecules in the chlorosomal rod elements, BChl a molecules in CsmA, FMO protein, and reaction center (the double diamond represents the P840 special pair), and the primary acceptor Chl aPD in the reaction center; cubes represent [4Fe-4S] clusters in the reaction center and soluble ferredoxin; rhomboids represent [2Fe-2S] clusters in CsmI, CsmJ, CsmX, and PetC; squares represent heme groups in the reaction center, PetB, and soluble cytochrome c; double hexagons represent menaquinone molecules in the reaction center and cytoplasmic membrane; triple hexagons represent a FAD cofactor.
3. The Oxygen Evolving Complex (OEC) is assimilated by Cyanobacteria (2.7 - 2.5 Ga)
The evolution of two Photosystems: The evolution of the oxygenic photosynthetic reaction center is currently under intense research scrutiny. There are two photosystems, known as Reaction Center Type I (RC1 or PSI)and Reaction Center Type II (RC II or PSII). The OEC became associated with RC II, but we find that both Photosystems are used by prokaryotes with slightly differing functions and evolutionary adaptations. Cyanobacteria use the oxygenic PSII and PSI electron transport system as does algae and plants. Anoxygenic bacteria use either RC I, or a prototype of RC II, depending on species and environment. At least one organism, Oscillatoria, has both.
To understand the various distribution and functions of the two reaction centers, and how they might be related evolutionarily, John F. Allen (Dept of Biology, Queen Mary, University of London) recently published a model to explain the appearance of the two reaction centers throughout the prokaryotes and higher plants and algae. It is now generally accepted that PSII evolved from a primitive PSI, the two reaction centers are in fact homologous in both function and structure (determined from kinetic, X-Ray and genetic studies). Dr. Allen calls his model a "redox switch hypothesis" in which the redox levels in the environment (amount of available reductants, etc) trigger a bacteria to switch from one reaction center to another. The associated light harvesting chlorophyll proteins, being integral within the bilipid membrane, are capable of serving as light harvesting complexes for both photosystems. Dr. Allen's model is presented below.
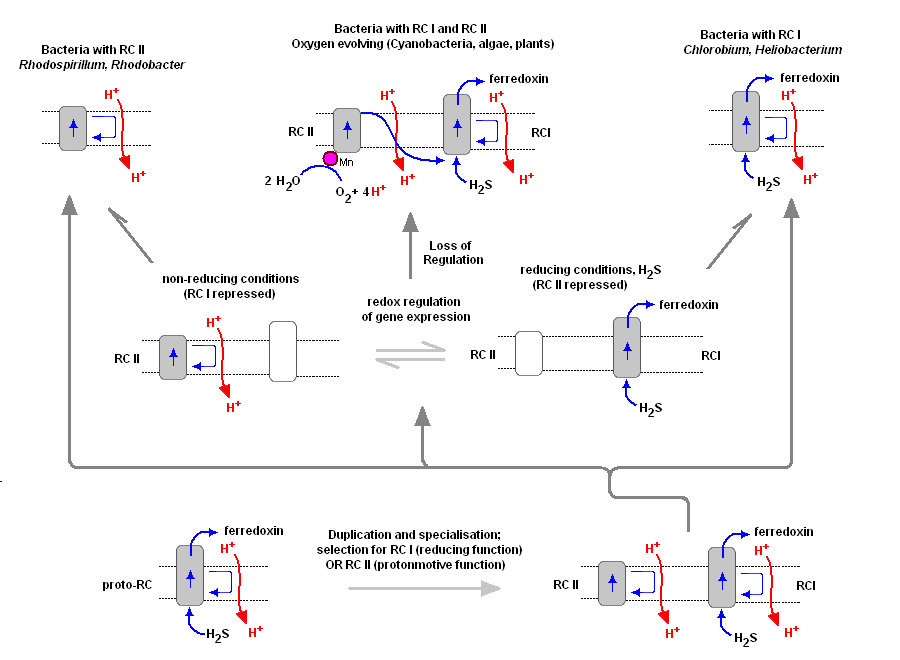
The original RC I is shown in the lower left, labeled as proto-RC. This reaction center operates with external reductants, such as hydrogen sulfide, and is capable of supplying electrons to a quinone pool, which can then recycle the energized electrons back to the reaction center while moving protons across the membrane into the periplasmic space thereby creating a protomotive force for the synthesis of ATP. In addition, the RC I provided electrons to soluble electron acceptors (ferredoxin) for use in organic synthesis.
Over time and changing conditions, a second independent photosynthetic reaction center emerged, under a genetic control scheme which would favor one over the other. Allen argues that in this early stage the two reaction centers would have been cooperative, their expression genetically controlled by changing environmental conditions (alternatively, there may have been initial competition). The rise of the RC II enabled the bacteria, under conditions in which a reductant, such as H2S, was readily available to support photon-driven proton pumping through the quinone cycle into the periplasmic space for ATP synthesis. Single reaction center anaerobic phototropohs were either photolithotrophic (RC 1) or photoorganotrophic (RC II). Dr. Allen argues that metabolic flexibility may have selected those bacterial capable of providing one or both of the two types of photosystems, depending on environmental conditions.
Over time bacteria appeared (such as in the present day Oscillatoria) in which both photosystems are present, and are synthesized and distributed through the membrane depending on environmental conditions encountered (Allen proposes this is carried out under redox control of gene expression). Oscillatoria limnetica has both RC I and RC II, and under conditions of low H2S, switches to RC II and performs oxygenic photosynthesis (Oscillatoria is a member of the Cyanobacteria. Note that there are other conditions, such as the heterocyst of Nostoc, in which RC II is shut off, as oxygen will interfere and shut down nitrogen fixation. See also C4 cell et al.).
As the image above shows, bacteria with RC II only are known (anoxygenic), as well as bacteria with RC I only (also anoxygenic) Dr. Allen proposes that at some point in time the genetic "redox switch" was not longer needed and the two photosystem began to specialize and work in cooperation. Genetic regulatory control can still take place by either lowering or increasing either the reaction centers themselves or distributing their associated light harvesting complexes between the two.
4. The molecular structure of the manganese-calcium oxide water splitting complex
Resolution: The recent 3.5 Angstrom resolution of the OEC has opened a black box that has been closed for many years. Earlier kinetic and spectroscopic data are falling in line with the mechanism of this complex but the race is on to determine exactly how the oxygen evolving complex works. We now know there are 4 manganese atoms (their respective redox states and behavior has not been determined with certainty), a calcium atom, now believed to be an essential component of the water oxidizing site, and a chloride (Cl-) co-factor. Chloride does not appear to be essential but is believed to be a charge stabilizing co-factor. An excellent animation of the Photosystem II complex, including the light harvesting chlorophylls, the reaction center and the OEC can be downloaded at Dr. Johannes Messinger's site (36 megs, Windows, RealTime) and is a beautiful rendition of PSII (highly recommended).
Dr. Messinger suggests that once the mechanism of water oxidation has been determined, it may be used to engineer an energy source by combining the oxidation of water with a hydrogenase (futuresolar hydrogen and oxygen production).
The mechanism of water oxidation has eluded scientists for decades. In the last 50 years much work has been carried out on the kinetics of PSII (OEC). In addition electrophoresis and related techniques carried out over the last 30 years had identified most of the polypeptides associated with PSII. New X-Ray diffraction studies have identified the stereochemistry and atomic structure of the OEC and surrounding complexes. It appears now that the race is on to determine the mechanism of water splitting (one of biology's greatest secrets). Once the mechanism of water oxidation is elucidated, it will become the greatest milestone of biochemistry since the discovery of DNA's structure in 1954.
 Photosystem II consists of an array of proteins, held together by numerous salt links, hydrogen bonds, water molecules et al. and can become especially
labile under stressful conditions such as high temperatures and low pH. The complex is also sensitive to photoinactivation if electron
flow throw the "Z" scheme is not delicately balanced (ATP phosphorylation of the surrounding light harvesting chlorophyll proteins
is part of this regulatory scheme (x)).
Photosystem II consists of an array of proteins, held together by numerous salt links, hydrogen bonds, water molecules et al. and can become especially
labile under stressful conditions such as high temperatures and low pH. The complex is also sensitive to photoinactivation if electron
flow throw the "Z" scheme is not delicately balanced (ATP phosphorylation of the surrounding light harvesting chlorophyll proteins
is part of this regulatory scheme (x)).
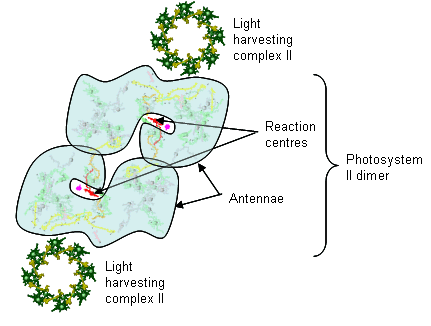
From the image at left, PSII is diagrammatically depicted as a dimer, as it is found embedded in the thylakoid membranes (see Dr. Messinger's video, which clearly shows the central axis of this complex). The PSII reaction center complex is surrounded by additional light harvesting chlorophylls, shown here as LHCII (only 2 are shown, these would completely surround the complex in a natural state.
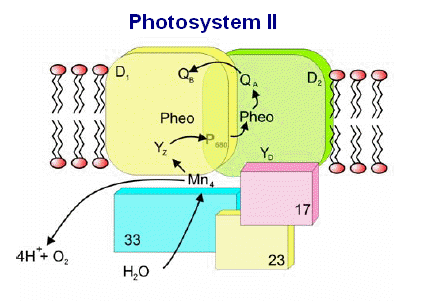 The
various proteins that surround the PSII complex are shown in the diagram at left. Then include the principle Light Harvesting Complexes
(LHCI), D1, and D2, as well as associated peptides including the 33 kdal, 23 kdal and 17 kdal. These periphery proteins support the
reaction center and associated cytochromes and other pigments.
The
various proteins that surround the PSII complex are shown in the diagram at left. Then include the principle Light Harvesting Complexes
(LHCI), D1, and D2, as well as associated peptides including the 33 kdal, 23 kdal and 17 kdal. These periphery proteins support the
reaction center and associated cytochromes and other pigments.
The general reaction is also outline (detailed below) in which the reaction center chlorophyll dimer, P680, is initially oxidized by an incoming photon, the electron being passed to the nearby pheophytin and then onto the quinone pool. Manganese, bound to water, calcium, chloride and oxygen (in manganese oxo bridges) then rapidly reduces Yz (a local Tyrosine) which itself became oxidized when it reduced the oxidized P680+. The basic process of water oxidation's mechanism was worked out 35 years ago by Joliot (X) and Kok (Y). Each PSII reaction center within the membrane works independently of one another and cycles through a set of 5 oxidation states, known as the "S" states (So-> S4). Our current understanding of the mechanism of water oxidation proceeds through these four states known as the "Kok Scheme" (for more details on the structure of PSII proper see the references and links provided below.
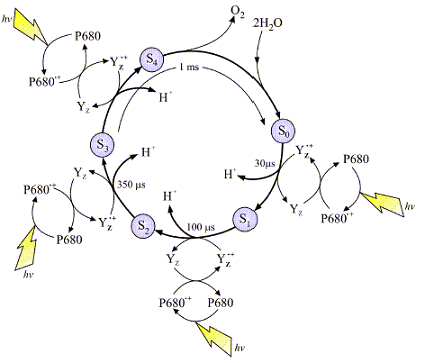 The
image at left is a typical representation of the Joliot-Kok Cycle. This particular schematic is from an article published by Dr.
James Barber, Imperial College of London, International Journal of Photoenergy 6:43-51 (2004). Make sure you take the time to visit Dr. Barber's dedicated web site to see his
Photosystem II images, they are very good.
The
image at left is a typical representation of the Joliot-Kok Cycle. This particular schematic is from an article published by Dr.
James Barber, Imperial College of London, International Journal of Photoenergy 6:43-51 (2004). Make sure you take the time to visit Dr. Barber's dedicated web site to see his
Photosystem II images, they are very good.
The OEC cycles according the cycle at left and requires four photons to complete the process. The OEC starts at State So, and then with each incoming photon moves on to Sate S1, then S2, S3 and finally S4. There are a total of five oxidation states of the OEC during this process. As the OEC cycles through this process four electrons are removed from bound water (see mechanism below). The deposition of protons during this process varies but it is believed 2 protons are released during the final S3 -> S4 transition. The schematic at least depicts one proton released per photon absorbed. Yz is a nearby Tyrosine residue, and P680 the reaction center special chlorophyll dimer (as discussed).
The X-Ray data and model of the OEC are shown below.
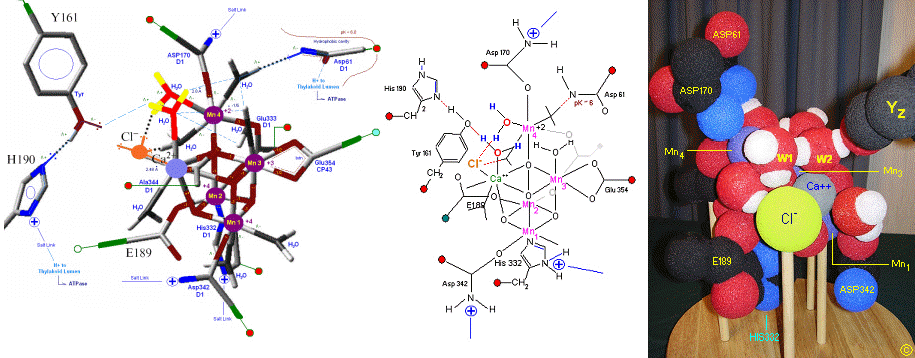
The original X-Ray at a resolution of 3.5 Angstroms, was reported by Ferreira et al in 2004. The image to the left, above, was taken from an article published by James P. McEvoy, Yale University (X). The image was colored coded and the middle image drawn directly from that. The model on the right was built using this X-Ray information.
The salient features of the complex (not all natural ligands may be in place, and McEvoy added an additional water ligand to complete the complex) include the manganese-calcium oxo bridging of the complex. The manganese atoms are labelled 1, 2, 3 and 4, with manganese number 4 protruding outside the main cubical structure below it, which includes manganese oxo binding, between the Mn 1, Mn 2 and Mn 3. Calcium is placed as one of the cuboidal corners (shown in blue on the left). Chloride is believed to reside on the outside of this cubane complex, and in our model on the right, appears to sit nicely (electrostatically) amongst the water molecules and calcium that surround it.
Yz, the primary electron acceptor of the OEC, sits to the left, approximately 3.4 Angstroms away from the chloride. Note the numerous ligands supporting the complex, which includes several Aspartates (contributed from the nearby D1 protein) as well as a histidine residue (near the bottom), also from D1. The substrate water molecules, labeled W1 and W2 in our model (right) is, at this time, an arbitrary assignment since the exact details of the mechanism is not yet known (we chose these water molecules after the model was built because of their proximity to Mn 4, and to illustrate a potential source of oxygen in our mechanistic scheme below. [when this article is complete tomorrow am (6-2) these images will be clickable to yield a closer view]. A proposed water channel may be directed out from aspartate 170 (D1) since it is believed that Tyrosine and histidine (left of the manganese cube) reside in a hydrophobic environment. See the reference for Dr. Johannes Messinger below for a review of currently proposed mechanism for water oxidation.
Water oxidizing Mechanism (proposed from a physical model built by the author): The following mechanism of water oxidation below is derived simply from the model above, and a few tips from the literature (nothing any student after 1 semester of Biochemistry could not propose). It is presented here to encourage biochemistry students to take the time, when they encounter reactive sites, especially those with unknown mechanisms, to read the literature and "give it a shot". Of course, realistically the chemistry must fit the known spectroscopic data, electron transfer permissions, and much in the end the mechanism may be very complex. But if one proposes a model, and follows the data, you might find yourself more involved in the determination of this event. (Note that the physical model was built directly from the 2001 X-Ray image).
The model makes the following assumptions (again, having limited time to read the literature, a few arguments were pulled out of the literature that seemed to match the physical model and then a mechanism was sketched out and is presented here). Some of the leading models now being proposed may be found in Johannes Messinger's review article, cited below, entitled Evaluation of different mechanistic proposals for water oxidation in photosynthesis on the basis of Mn4OxCa structures for the catalytic site and spectroscopic data, Phys. Chem. Chem. Phys., 6:4764-4771 (2004).
The literature may have advanced beyond the mechanisms proposed in Messinger's review, and one should search them out in order to follow the course of the OEC's mechanistic discovery (the mechanism of water oxidation will be one of the milestones of biochemistry (in this author's opinion, the greatest discovery since DNA in 1954).
For the purposes of this exercise the following assumptions are made: (1) only 1 Manganese is redox active (Mn number 4 - most models involve at least two manganese and base their argument on the oxidative potential necessary to carry out the final step of water oxidation); (2) Tyrosine 161 (Yz) removes protons from the reaction center at every step through the Kok S-state cycle (note that the proton release pattern, not shown in the Kok diagram above, is believed to be 1 proton from So -> S1, no protons from S1 -> S2, 1 proton from S2 -> S3, and 2 protons from S3 -> S4; (3) there is an attack on a Mn=0 oxo linkage; [Note: although not reported, one might envision a Mn triple bond with oxygen - at least one metallo-oxygen evolving scheme uses this] (4) a peroxide is formed at the S3 state; (5) one substrate water forms a ligand to Ca++ (6) one substrate water is forms a ligand to Mn 4 (5 and 6 based simply at looking at the physical model. Chloride is assumed to play a co-factor supporting role.
The above assumptions, and using the physical model above (Styrofoam balls from Wal-Mart) as a practical guide to stereochemistry; the following mechanism is proposed. Recent X-Ray data suggests that protons are removed (via a water channel) on the Asp 61 side of the OEC (the argument being that Yz (Y161) is in a hydrophobic pocket and removed from what appears to be a channel to the lumen on the Asp 61 side of the complex). Since the mechanism proposed below requires Yz to extract protons with incoming photons, the protons are modeled here as being removed on the Yz side of the OEC complex (its just a model!).
State So: Resting state. The two substrate water are colored. Mn 4 begins in a +2 state. Mn 3, Mn 2 and Mn 1 are in a +3, +4, +4 state (and remain that way being redox inactive for this scheme). Only Mn 1 is redox active. Protons are numbered 1-4. Note that the two substrate water molecules were chosen simply based on their suggestive positions from the physical model above. No argument is made here as to D1 ligand changes (for example, Mn in its lower oxidation state may prefer a hexagonal arrangement and may move to pentagonal coordination as its oxidation step increases but this is not considered here). Photon arrives at P680.
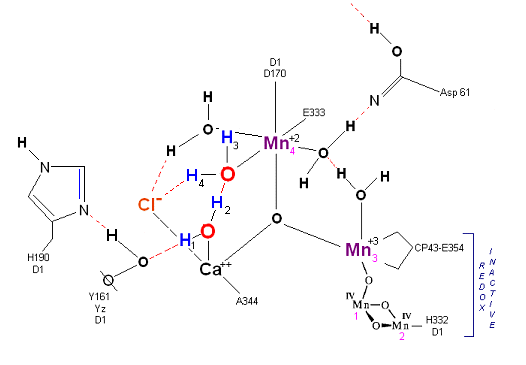
State So': First photon arrives. P680+ reduced by Yz. Initial + charged carried by hydrogen bridge between Mn 2 and Ca (note that in the physical model this transitory bridge would be linear). Cl- helps to stabilize the positive charge. Notice that in a one proton extraction scenario such as this Histidine 190 may spin 180 degrees before resting in the S1 state (alternatively a simple 2e- resonance action could result in relaxation to S1).
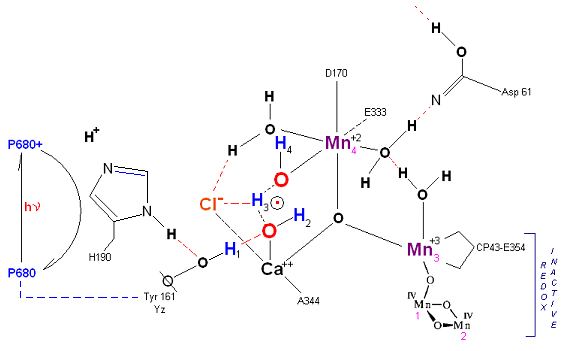
State S1: S1 resting state. H3 transfers from substrate H2O #2 to H2O #1. Redox reactive Mn 4 is now in the Mn(III) oxidation state. Next photon arrives at P680.
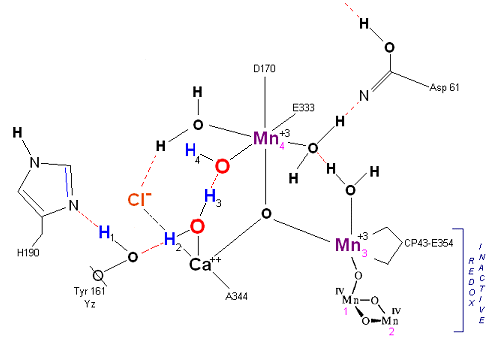
State S1': P680+ is reduced by Yz. Hydrogen bridge forms as in So -> S1, charged stabilized by Cl-.
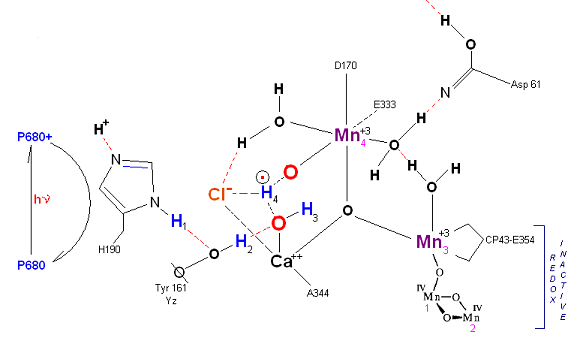
State S2: S2 resting state. Mn 4 is now an oxo-manganese (carbonyl) as Mn(IV), with a double bond to the original substrate H20 #2. Ligands around Mn 4 may shift here. Cl- still serving in a stabilizing fashion. Note that the Tyrosine-Histidine system is moving protons out one at a time. Next photon arrives at P680.
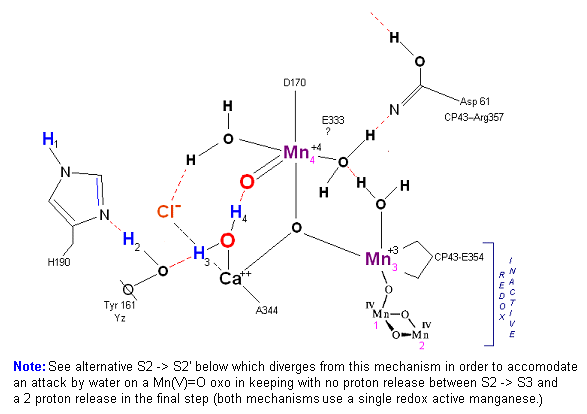
State S2': P680+ is reduced. This time the intermediate is a peroxo-radicle. The move to the resting S3 state involves the reduction of Mn 4 from Mn(IV) to Mn(III) [Note: in this particular mechanism Mn never moves to Mn(V) which is required in several models to achieve a high enough oxidation potential to complete the Kok cycle.] It is possible that H4 may bridge the two oxygens to stabilize the radical.
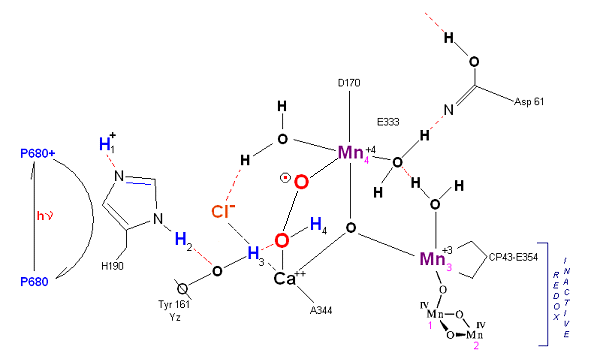
State S3: S3 resting state is a stable peroxide. Mn 4 is now back to a Mn(III) oxidation state. Only one of the orignal Hydrogens remained bound. Next photon arrives at P680.
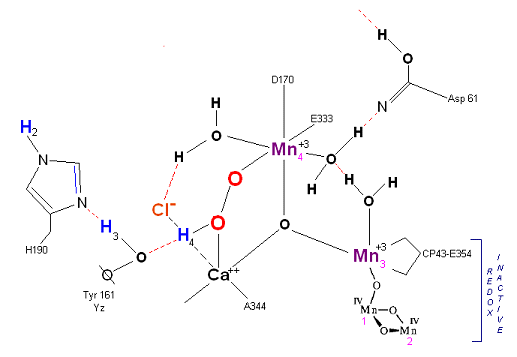
State S3' --> S4: Finally in a series of 1 electron transfers, following the reduction of P680, Mn(III) returns to its ground state Mn(II), oxygen is formed, and the system prepares itself to be reset to the initial State So.
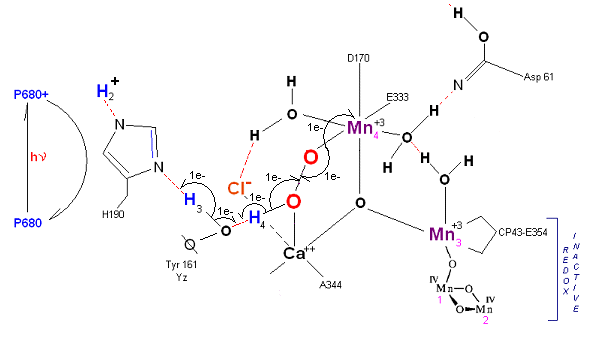
State S4 --> S4':
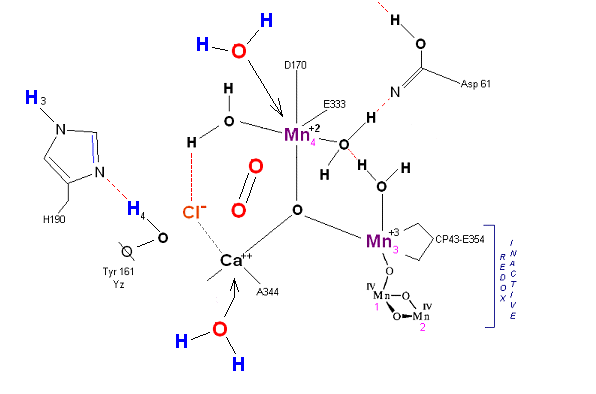
Incoming water molecules resets the OEC to State So. The point of this exercise was to demonstrate to undergraduate students that starting from simple models, and reading the literature, one can at least make an attempt to proceed through an enzymatic reaction using simple electron transfer principles. The above mechanism could have been devised by a first semester biochemistry student. It does not portend to replace the serious work being carried out at this time in researching the actual mechanism, but only demonstrates to the student that by proposing (after reading the literature) a simple mechanism one brings the subject matter closer to an understanding.
1.Lowe, D. R., Early Life on Earth, Nobel Symposium No. 84, ed., Bengtson, S., Columbia Univ. Press, New York, 24-35 (1994)
2. Paul Preuss, Science Beat Spinach, Or The Search For The Secret Of Life,
July 2004.
3. Michael Russell, First
Life, The American Scientist Volume: 94 Number: 1
Page: 32 (American Scientist Online)
4. Johannes Messinger, Evaluation of different mechanistic proposals for water oxidation in photosynthesis on the
basis of Mn4OxCa. Physical Chemistry Chemical Physics, 2004, 6,4764-71.
4. Russell, M. J., Daniel, R. M., Hall, A. J., Sherringham, J. A., J. Mol. Evol., 39:231-43 (1994)
5. Anbar, A. D., Holland, H. D. Geochim. Cosmochim. Acta, 56:2595-2603 (1992)
6. Dawson, Scott, Creatures
from the Black Lagoon: Lessons in the Diversity and Evolution of Eukaryotes, Berkeley University, 1-18
7. Brandes, Jay and Hazen, Bob, Key chemical in life creation - ammonia -
created at Hydrothermal Vents. Carnegie Institution's Geophysical Laboratory (1998)Rasmussen, Birger, Filamentous microfossils in a 3,245-million-year-old
volcanogenic massive sulphide deposit. Nature 405 676-679 (June 2000), Dept. of Geology and Geophysics, University of
Western Australia, Nedlands, Western Australia 6907, Australia
8. Rasmussen, Filamentous microfossils in a 3,235-million-year-old sulphide deposit
9. Blank, Biologist's Find Alters The bacteria Family Tree, Science
Daily, Department of Earth & Planetary Sciences at Washington University in St. Louis
Photosystem II - General references
1. Barber, James, Towards a full understanding of water splitting in photosynthesis, International Journal of Photoenergy
6:43-51 (2004)
Have a comment? We are creating a "Teaching
with Molecular Models Weblog" (www link depository) The Weblog is under construction and should be ready by this fall (2006). Drop by!
2. Ferreira, Kristina, Iverson, Tina, Maghlaoui, Karim, Barber, James,
Architecture of the Photosynthetic Oxygen-Evolving Center, Science 303:1831-1837 (2004)
3. Govindjee and Krogmann, David,
Discoveries in oxygenic photosynthesis (1727-2003): a perspective, Photosynthesis Research 80:15-57 (2004)
4. Govindjee and Orr, Larry, Photosynthesis on the Web, Departments of Biochemistry and Plant Biology and Center
of Biophysics & Computational Biology, University of Illinois, Urbana, IL and Center for the Study of Early Events in Photosynthesis,
Arizona State University respectively (2005)
5. Govindjee, J. T. Beatty, H. Gest, and J. F. Allen, eds., Advances in Photosynthesis and Respiration Vol. 20: Discoveries in Photosynthesis,,
Springer, Dordrecht (2005). pp. 119-129
6. Govindjee, J. T. Beatty, and H. Gest eds., special issue: Celebrating the Millennium—Historical Highlights of
Photosynthesis Research, Part 2, Photosynthesis Research 76:93-103
7. Crofts, Anthony, Structure and Function in Photosystem II, Crofts Laboratory, University of Illinois
8. Haumann, Micahel, and Junge, Wolfgang,
Photosynthetic Water Oxidation: A Simplex-Scheme of its Partial Reactions, Biochem. Biophys. Acta 1411:1-6 (1999)
9. Hillier, Warwick and Wydrzynski, Tom,
The Affinities for the Two Substrate Water Binding Sites in the O2 Evolving
Complex of Photosystem II Vary Independently during S-State Turnover, Biochemistry 200, 39:4399-4405 (2000)
10. J.P. and Shock, E.L., Energetics of overall metabolic reactions in
thermophilic and hyperthermophilic Archaea and bacteria. FEMS Microbiology Reviews 25:175-243 (2001)
11. Kok, B., B. Forbush, and M. McGloin. 1970. Cooperation of charges in photosynthetic
oxygen evolution. I. A linear four step mechanism. Photochem. Photobiol. 11:467–475.
12. McEvoy, James P., Gascon, Jose A. et al.,
Computational Structural Model of the Oxygen Evolving Complex in Photosystem II: Complete Ligation by Protein, Water and Chloride, Photosynthesis:
Fundamental Aspects ot Global Perspectives, A. van der Est and D. Bruce, Eds., 278-80 (2005)
13. Messinger, Johannes,
Evaluation of different mechanistic proposals for water oxidation in photosynthesis on the basis of Mn4OxCa structures for the catalytic site and
spectroscopic data, Phys. Chem. 6:4764-4771 (2004)
14. Zouni, Athina, Horst-Tobias Witt, Kern, Jan et al.
Crystal structure of Photosystem II from Synechococcus elongatus at 3.8 Angstrom resolution, Nature 409:739-743 (2001)
15. Homann, P. H., Hydrogen metabolism of green
algae. Discovery and early research - a tribute to Hans Gaffron and his coworkers. Invited contribution. (2003)
16. ASU Center for the Study of Early Events in Photosynthesis,
Photosynthesis Center, AZ State Univ.
17. Des Marais, David J., Evolution:
When Did Photosynthesis Emerge on Earth?, Science, NASA Ames Research Center, Moffett Field, CA
![]()
![]()