
|
UBIQUITIN |

|
General |
|
Basic information PDB accesion codes [33] and 3D structures of human ubiquitin [34]: 1ubq (crystal structure of Ub, source: solid-phase chemical synthesis); 1ubi (crystal structure of Ub, source: human erythrocytes); 1d3z (NMR structure of Ub in agueous buffer, source: human, expressed in E.coli); 1aar (crystal srtucture of Ub2, source: solid-phase chemical synthesis); 1tbe (crystal structure of Ub4, source: human); 1f9j (crystal structure of Ub4, source: human, expressed in E.coli). Genes and cellular expression: There are two main structural arrangements of ubiquitin gene. The first arrangement is the polyubiquitin gene, consisting of tandem repeats of an ubiquitin coding sequence. Number of repeats varies depending on species and can be different even within one species. Ubiquitin is synthesized as a precursor polyubiquitin protein, which is characterized by „head” to „tail” repeats [27]. In some species there is an amino acid after the last repeat, Tyr in chicken, Val in human [34]. Mono-ubiquitin is post-translationally cleavaged from the primary translation product. The second arrangement of ubiquitin genes consists of single ubiquitin moiety fused to an unrelated protein. Expression of this gene results in a fusion protein consisting of one ubiquitin at the N-terminus and ribosomal protein L40 at C-terminus. Also S27a ribosomal protein is synthesized as a fusion protein [34]. Also S30 ribosomal protein is synthesized as fusion protein with the ubiquitin-like protein fubi [38]. There are multiple ubiquitin genes in human genome, many of them are non-functional reserve-transcribed pseudogenes. There are three types of mRNA generated by transcriptionally active genes: UBA, UBB and UBC with the number of nucleotides approximately 600, 1000 and 2500, respectively. The UBA transcripts belong to the second arrangement type of ubiquitin genes and encode human ubiquitin-ribosomal fusion protein and some other fusion proteins. Stability and Folding: Ubiquitin is a very stable protein and is characterized by significant resistance against extreme values of pH, temperature and proteolysis [2,11,35]. Only two terminal glycines are often lost during proteolysis process. Ubiquitin owes its stability mostly to abundant hydrogen bonds, there are not disulphide bonds, coordinated metal ions or binding cofactors [13,18,36]. Investigations showed that ubiquitin folds in a simple two-state process in an aqueous solution (N <-> U, where N, U correspond to native and unfolded state, respectively) [12,14]. In aqueous methanol a partial unfolded state (A) of ubiquitin is observed (N <-> A). It is identified that N-terminal fragment (residues 1-35) prevents native structure, which corresponds to the beta-hairpin (residues 1-17), and the hairpin-helix fragment of ubiquitin (residues1-35) [17]. It is worth mentioning that the peptide corresponding to the N-terminal beta-hairpin of ubiquitin was also found to form some residual structure in aqueous solution [16]. Also a very interesting is that two unfolded polypeptide chains, one corresponding to 1-35 and the second to 36-76 residue sequence of ubiquitin, were found to fold the same native structure as ubiquitin in aqueous solutions. Two polypeptides mentioned above mixed in the molar ratio of 1 : 1 are, according to NMR data, in equilibrium with the reconstructed native structure [15,17]. Subcellular location: Ubiquitin is present in cellular cytosol and nucleus [34].
|
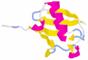





|
SCOP classification [33]: |
|
|
Class: |
alpha and beta proteins (a+b) |
|
Fold: |
β-Grasp |
|
Family: |
ubiquitin-related |
|
Superfamily: |
ubiquitin-like |
|
Protein: |
ubiquitin (Ub) |
|
Species: |
Homo Sapiens |