|
UBIQUITIN |

|
Structure |
|
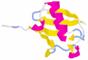
Tertiary structure: (this describes how the secondary structure elements are arranged to form the overall holding pattern of the polypeptide) The three-dimensional structures of ubiquitin and its multi-ubiquitin chains were investagated and solved by NMR (Nuclear Magneic Resonance) - PDB: 1d3z; and X-ray crystallography - PDB: 1ubi, 1ubq, 1aar, 1tbe, 1f9j [1,9,33]. Figure 2. PDB structure of 1d3z. Figure 3. PDB structure of 1ubi. Figure 4. PDB structure of 1ubq. Figure 5. PDB structure of 1aar. Figure 6. PDB structure of 1tbe. Figure 7. PDB structure of 1f9j. Figures 2-7 present three-dimensional structures of human mono- and multi-ubiquitins with exposed secondary motifs. Yellow fragments correspond to β-strands, red ones to helices, sea-blue characters are amino acids that make up β-turns and white-gray ones correspond to non-motif fragments. 3D structure of ubiquitin is very stable and the differences between forms of ubiquitin in water solution and crystalline forms are nearly not noticeable (Fig. 2,3,4). Interesting from the biological point of view is the 3D structure of multi-ubiquitin chains. These structures are not easy to predict on the basis of the knowledge about 3D structure of the mono-ubiquitin molecule and features of isopeptidic bond between ubiquitins that make up the chain. Different values of ψ and Ф angles of C-terminal glycines and variety of rotation angles of methylene groups of lysine (48 or 11) causes big diversity of structures which can appear. Some progress in this matter was achieved thanks to the investigations on the structure of crystalline di-ubiquitin (Fig. 5). Di-ubiquitin chain is characterized by: a). Connection of ubiquitins by isopeptidic bond created between Gly76 of one ubiquitin and Lys48 of the second molecule; b). Hydrophobic residue stretch placed on the ubiquitin surface and related to the same amino acid residues (Leu8, Ile44, Val70) on the second ubiquitin abort the axis of symmetry to form hydrophobic pocket; c). Hydrogen bonds created by two ubiquitin moieties with the residues of Gly47, Gln49 and Leu71. Some more informations about the structure of poly-ubiquitin chains was given by tetra-ubiquitin X-ray crystallographical data (Fig. 6). Only co-ordinates for first and second ubiquitin were provided by the crystal structure of Ub4. The remaining co-ordinates of third and fourth moieties were generated by translation of 24.1 Ǻ along the a-axis of the molecule. Tetra-ubiquitin chain is characterized by: a). β-Sheets in ubiquitin first and third ones differ from native mono-ubiquitin, the first two strands are shorter; b). 2-fold screw axis of symmetry, while di-ubiquitin has simple 2-fold axis of symmetry. More recently a novel isomer of tetra-ubiquitin was found and characterized by X-ray crystallography (Fig. 7). |