
|
UBIQUITIN |

|
Ubiquitinylation |
|
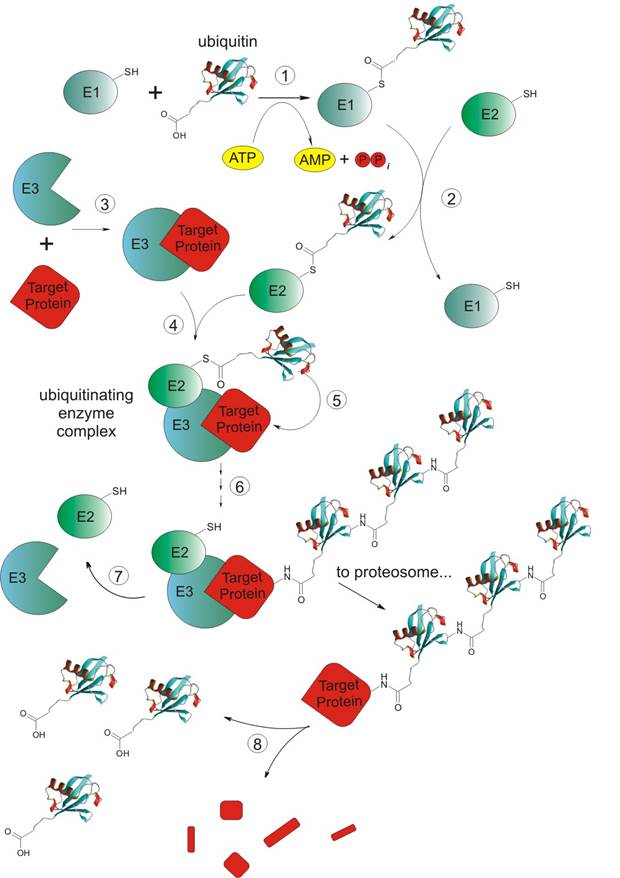
Introduction: In the early 70’s it was realized that proteins were continually synthesized and degraded in cells, and that some proteins turned over more rapidly than others. But the accurate way of the breakdown of proteins back to amino acids in the cells was not completely understood. The lysosome system of mammalian cells was known to degrade intracellular proteins but later scientists noticed that protein degradation occurred in rabbit reticulocytes (the precursors of erythrocytes) [6] which lack lysosomes. That was the way how rabbit reticulocytes became model cells to study non-lysosomal intracellular proteolysis. A cytosolic proteolytic system in reticulocytes was found, surprisingly, to be dependent on energy taken from ATP hydrolysis [6,8]. Fractionation of reticulocyte cytosol generated two fractions (A and B) which were required for at least some of the ATP-dependent proteolysis of some test proteins. The active factor in A fraction was purified, called APF-1 (ATP-dependent Proteolysis Factor-1), and found to be covalently conjugated to proteins in the presence of ATP and B fraction. APF-1 was then shown to be identical to ubiquitin [8]. Proteins targeted by ubiquitin [39]: a). Cell surface receptors; b). Tumor suppressors and growth modulators; c). Transcriptional activators and inhibitors; d). Cell cycle regulators (e.g. the cyclins); e). Mutant and damaged proteins. Steps of ubiquitinylation (or ubiquitylation): In general non-lysosomal ubiquitin dependent degradation is based on attaching C-terminus of ubiquitin to e-amino group of Lys of target protein creating isopeptidic bond. Ubiquitin dependent degradation path-way consists of four enzymes systems (E1: ubiquitin-activating enzyme, E2: ubiquitin-conjugating enzyme, E3: ubiquitin-ligating enzyme, and E4(?) only suspected to exist) and 8 steps [7,23]: Figure 8. Scheme of ubiquitinylation process (description: see below). 1). Activation process. C-terminal residue of ubiquitin (exactly a product of the reaction between ATP and ubiquitin at the presence of Mg2+ ions - ubiquitin adenylate) is covalently (-) attached to a Cys sulphydryl residue in E1 enzyme. Prodcts are: ubi-COS-E1, AMP and PPi [41]; 2). Transfer of ubiquitin on Cys sulphydryl residue of E2 (exactly on the residue of E25) enzyme. Products are: ubi-COS-E2, E1; 3). Recognition of targeting protein by E3 enzyme and creation of non-covalent complex between E3 and protein designed for degradation. Product is: E3--Protein; 4). Obtaining an ubiquitinating enzyme complex by creation of a non-covalent (--) bond between non-covalent complex created previously and E2 enzyme. Product is: ubi-COS-E2--E3--Protein; 5). Transfer of ubiquitin from E2 to the target protein Product is: E2--E3--Protein-NHCO-ubi; 6). Repetition of ubiquitin transfer many times. Product is: Protein-NHCO-(ubi)n, where n>1. Ubiquitins are attached to each other by C-terminus of one ubiquitin and Lys48 (also Lys11) of another one. Some scientists claim that tetra-ubiquitin (Ub4) is the minimum ubiquitin chain required for recognition of ubiquitin chains by the S5a sub-unit of 16S proteosome; 7). After the dissociations of E2 and E3 enzymes target protein is taken to the proteolysis (performed by 26S proteosom complex [42]). Products are (after dissociation): Protein-NHCO-(ubi)n, E1, E2; 8). Releasing of the ubiquitin performed probably by protease 26S and its reuse. Main enzymes involved in ubiquitinylation process: a). E1: only one known, consists of one or two polypeptidic chains, with M.W. 105 kDa per chain, very conservative protein; b). E2: 5-12 proteins of this type known (depending on literature), homologous family, e.g. E25 carries ubiquitin in the presence of E23, E25 participates in DNA reparation process; c). E3: many and structurally unrelated, characterized by ultimate biological specificity, M.W. about 250 kDa, very hard to analyze; d). E4: not known yet, but postulated as existing [37]. Varshavsky’s law[21,22,37]: Every cellular protein has its own half-life time, which depends on its role in living cell. Half-life times have different lengths for different proteins (see table below). Table 1. Chosen half-life times of human and rat proteins. There is a very important question connected with half-life time of the protein. The point is to know what feature of protein has the biggest influence on the length of its half-life time. According to Varshavsky’s group investigations the half-life time is determined by N–terminal amino acidic residue of protein. They performed very interesting experiment which result was the confirmation of this hypothesis. They expressed in E.coli bacteria chimerical protein ubi-Met-β- galactosidase, where ubiquitin was connected to N-terminal methionine residue of β-galactosidase by its own C-terminal glycine residue. Next yeast was used for this protein expression and only β-galactosidase was obtained. This proved the presence of proteolitic enzyme system in eukaryotic cells, which degrades chimerical protein. Later Varshavsky decided to express some other chimerical proteins ubi-X-β-galactosidase, where X corresponded to different amino acid residues as a N-terminus of β-galactosidase. For new expressed chimers different half-life times were noticed. The longest half-life times belonged to X equal to Val, Ser, Cys, Met, Ala, Thr, Gly (over 20 hours). The medium half-life times were given by residues like Ile and Glu (half of an hour) Tyr, Gln, His about 10 minutes, but the shortest belonged to Leu, Phe, Trp, Asp, Asn (about 3 minutes) and Arg about 2 minutes. So by modifying of N-terminus we can regulate half-life time of target proteins for ubiquitinylation. During next few years many proteins were investigated in the same way and these investigations confirmed Varshavsky’s N-terminal rule. Short summary: The enzymatic pathway that conjugates ubiquitin to proteins was elucidated and a protease that degrades ubiquitinylated proteins was idntified and characterized as the 26S proteosome. Also many of the proteins which are targets for ubiquitin-mediated degradation have been identified. It is suspected that the ubiquitin-mediated proteolytic system is probably responsible for the degradation of most short-living proteins. The ubiquitin system selectively recognises proteins via some features of their structure, which in turn may be regulated e.g. by phosphorylation or allosteric interactions. Some novel investigations show that ubiquitin function is not only the regulation of protein turnover but also that ubiquitinylation might serve to modify some proteins, and therefore their activity (monoubiquitinylation of histone H2A), rather than triggering their complete breakdown to amino acids [38]. |





|
Tissue or organ |
Organism |
Half-life time [days] |
|
Total protein amount of organism |
Human |
80 |
|
Rat |
17 |
|
|
Proteins of liver and plasma |
Human |
10 |
|
Rat |
6 |
|
|
Proteins of mussles, skin and skeleton |
Human |
158 |
|
Rat |
21 |
|
|
Proteins of mussles |
Human |
180 |
|
Rat |
24-30 |