Dr. Marc van der Kamp
My research interests centre on enzyme specificity, catalysis and dynamics and the effect of mutations on protein dynamics and stability. These interests started in my years as an undergraduate at Wageningen University. During two MSc research projects, I gained experience with both computational and experimental biochemical methods. I subsequently specialized in computational modelling during an internship at a pharmaceutical company and a pre-doctoral research position, where I developed protocols to predict enzyme-substrate binding. Thereafter, I came to the Mulholland group in Bristol as a Marie Curie Early Stage Research Fellow.
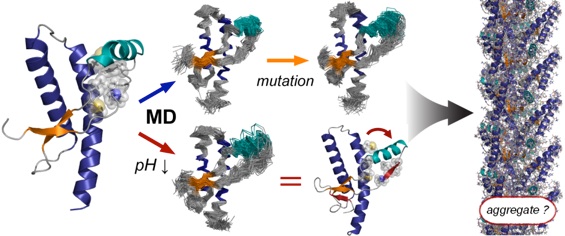
After obtaining my my PhD, I went to the Daggett group at the University of Washington (Seattle) for about 2 1/2 years of post-doctoral research. I gained extensive experience with performing and analyzing molecular dynamics simulations of proteins. I primarily focussed on the effect of the environment and mutations on the dynamics and stability of the prion protein, leading to insights into pH- and mutation-induced instability involved in causing disease.4,6,8 I further contributed to the creation and use of a database with simulations to investigate protein dynamics and folding across 'protein fold space' (www.dynameomics.org)3,7.
Currently, I am back in the Mulholland group working as a Post-Doc on a variety of projects (often in collaboration with experimentalists) related to enzyme dynamics, catalysis and design, again using QM/MM and molecular dynamics methods. My research interests further include other biomolecular modelling methods (homology modelling, docking, QSAR modelling and pure QM calculations), related experimental biophysical methods (e.g. protein crystallography and NMR), as well as the broader fields of biochemistry and chemical biology.
Publications
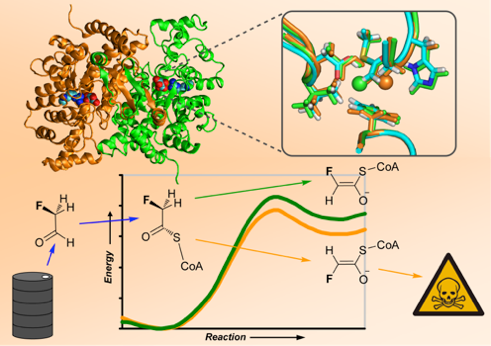
(1) Van der Kamp, M. W., McGeagh, J. D., and Mulholland, A. J. (2011) "Lethal synthesis" of fluorocitrate by citrate synthase explained through QM/MM modeling, Angewandte Chemie International Edition 50, 10349-10351.
(2) Van der Kamp, M. W., and Daggett, V. (2011) Molecular dynamics as an approach to study prion protein misfolding and the effect of pathogenic mutations, In Prion Proteins (Tatzelt, J., Ed.), pp 169-197, Springer Berlin / Heidelberg.
(3) Jonsson, A. L., Schaeffer, R. D., Van der Kamp, M. W., and Daggett, V. (2011) Dynameomics: protein dynamics and unfolding across fold space, BioMolecular Concepts 1, 335-344.
(4) Chen, W., Van der Kamp, M. W., and Daggett, V. (2010) Diverse effects on the native b-sheet of the human prion protein due to disease-associated mutations, Biochemistry 49, 9874-9881.
(5) Van der Kamp, M. W., Zurek, J., Manby, F. R., Harvey, J. N., and Mulholland, A. J. (2010) Testing high-level QM/MM methods for modeling enzyme reactions: acetyl-CoA deprotonation in citrate synthase, Journal of Physical Chemistry B 114, 11303-11314.
(6) Van der Kamp, M. W., and Daggett, V. (2010) Pathogenic mutations in the hydrophobic core of the human prion protein can promote structural instability and misfolding, Journal of Molecular Biology 404, 732-748.
(7) Van der Kamp, M. W., Schaeffer, R. D., Jonsson, A. L., Scouras, A. D., Simms, A. M., Toofanny, R. D., Benson, N. C., Anderson, P. C., Merkley, E. D., Rysavy, S., Bromley, D., Beck, D. A. C., and Daggett, V. (2010) Dynameomics: A comprehensive database of protein dynamics, Structure 18, 423-435.
(8) Van der Kamp, M. W., and Daggett, V. (2010) The influence of pH on the human prion protein: Insights into the early steps of misfolding, Biophysical Journal 99, 2289-2298.
(9) Van der Kamp, M. W., and Daggett, V. (2009) The consequences of pathogenic mutations to the human prion protein, Protein Engineering Design & Selection 22, 461-468.
(10) Van der Kamp, M. W., Shaw, K. E., Woods, C. J., and Mulholland, A. J. (2008) Biomolecular simulation and modelling: status, progress and prospects, Journal of the Royal Society Interface 5, S173-S190.
(11) Van der Kamp, M. W., Perruccio, F., and Mulholland, A. J. (2008) High-level QM/MM modelling predicts an arginine as the acid in the condensation reaction catalysed by citrate synthase, Chemical Communications, 1874-1876.
(12) Van der Kamp, M. W., and Mulholland, A. J. (2008) Computational enzymology: insight into biological catalysts from modelling, Natural Product Reports 25, 1001-1014.
(13) Van der Kamp, M. W., Perruccio, F., and Mulholland, A. J. (2007) Ab initio QM/MM modelling of acetyl-CoA deprotonation in the enzyme citrate synthase, Journal of Molecular Graphics & Modelling 26, 596-601.
(14) Van der Kamp, M. W., Perruccio, F., and Mulholland, A. J. (2007) Substrate polarization in enzyme catalysis: QM/MM analysis of the effect of oxaloacetate polarization on acetyl-CoA enolization in citrate synthase, Proteins: Structure, Function and Bioinformatics 69, 521-535.
(15) Rodriguez, A., Oliva, C., Gonzalez, M., van der Kamp, M., and Mulholland, A. J. (2007) Comparison of different quantum mechanical/molecular mechanics boundary treatments in the reaction of the hepatitis C virus NS3 protease with the NS5A/5B substrate, Journal of Physical Chemistry B 111, 12909-12915.

My PhD research (extended with an IBM PhD Fellowship) focused on the use of hybrid quantum mechanical / molecular mechanical (QM/MM) methods to investigate enzyme catalysis and specificity. I used both fast semi-empirical and state-of-the-art high-level QM/MM methods successfully to analyze sources of catalysis, mechanisms of reaction and stereospecificity.1,5,10-14 This work revealed previously unknown details about the reaction mechanism in citrate synthase, including the unsuspected role of an arginine acting as an acid11 and an explanation for the enzymatic specificity involved in 'lethal synthesis' of fluorocitrate.1