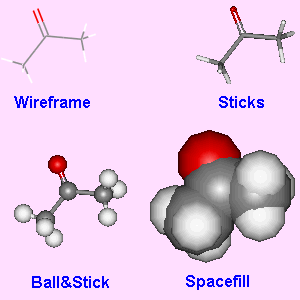
Fig.5. Examples of the four different display types for acetone.
There are many different packages that can be used to display 3D molecular structures. Some of these are stand alone packages which can be run without a browser, whereas others are browser plug-ins suitable for displaying 3D content inside web pages. However, before we compare the different packages, it is worthwhile looking briefly at the possible types of display that are available.
For small molecules, there are four main display types. Wire (or wireframe) is the most basic, with the atom positions just being joined by thin lines. Sticks (or cylinders) is similar to wireframe, except that the wires have now been thickened considerably. Ball&Stick represents each atom by a sphere of the same size, but with different colours for each different type of atom. The spheres are joined by sticks to represent the bonds. Spacefill represents each atom in a molecule with a sphere of relative size based on the van der Waals radius of the atom. Most 3D viewers allow easy switching between these 4 display types, and many of the parameters for each type (such as atom colour, diameter of atoms, etc.) can be user selected.

Fig.5. Examples of the four different display types for acetone.
For macromolecules, such as proteins, there are a few more display styles. Rather than display the whole molecule in, say, ball&stick style, which would be unneccesarily complex, the structures are often simplified by representing the primary shape of the structure, such as the alpha-helix, beta-sheets, or peptide backbone, by thick wires, ribbons or tubes. It is often possible with some packages to represent different parts of the molecule in different styles. So for example, to show the active site within an enzyme, the primary backbone of the molecule would be shown only as ribbons, but the atoms of interest would be highlighted by showing them in ball&stick style.
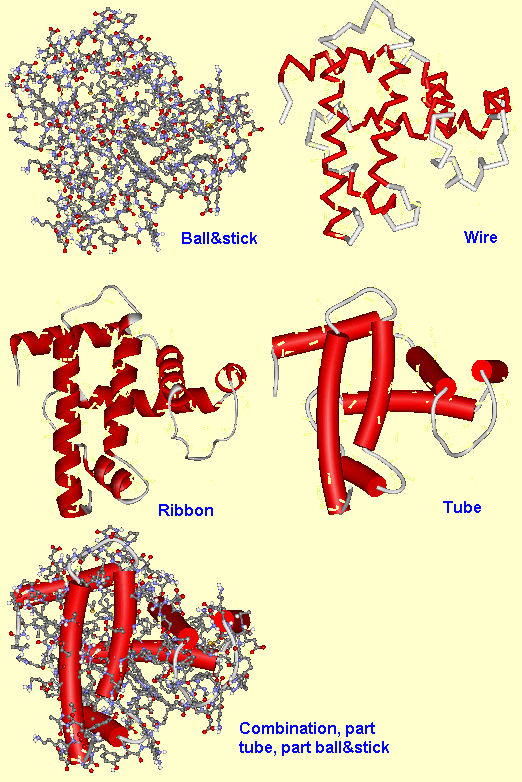
Fig.6. Examples of some of the extra display types for proteins.
(ii) Stand-alone packagesOne of the earliest packages that could show 3D molecular structures was Rasmol, and it is still popular today. Other packages which can be used to display structures include Chem3d, WebLab Viewer and HyperChem. Some of these allow 3D editing of the structures, as described in the previous section. All of these can be set up to act as external viewers to a web browser, so that when the browser tries to load a molecular structure file (with extension pdb, mol, xyz, etc.), the viewing package runs and displays the structure in a separate window. The structure files can then be saved to disk, or exported into different formats, for analysis off-line. Therefore, simply supplying the structure file with a published on-line paper provides the reader with a wealth of extra information that would not be possible in printed format. |
 Fig.7. A screen image of Rasmol displaying a spacefill structure of the anatoxin molecule. |
The disadvantage with using stand alone viewers as 'helper programs' is that the molecular structures appear in separate windows to the rest of the document. For this reason, a number of plug-ins have been developed for the most common web browsers (Netscape and Internet Explorer) which allow the 3D structure to be placed inside the existing web page. The mostly widely used on the web is called Chemscape Chime, which is based on the Rasmol engine and looks and behaves very similar to this. To insert a Chime structure into a web page, the author simply needs to add a few extra lines of HTML code detailing the name of the structure file and the display parameters. For example, the code needed to display the molfile for benzene would be:
<embed src="benzene.mol" width=200 height=200 display3d="ball&stick" startspin=true spiny=20 bgcolor="#FFFFAA" name="benzene">
where the parameter 'src' shows the name (and path) of the molfile, 'width' and 'height' determine the display area in pixels, 'display3d' defines the display type, 'startspin=true' starts the molecule rotating immediately the page is loaded, with rotation speed around the y direction given by 'spiny'. Finally the background colour 'bgcolor' is given in three pairs of hexadecimal numbers corresponding to red, green and blue. The 'name' parameter gives each structure on the page a unique identifier, which is needed for Chime scripts (see later). A screenshot of the resulting interactive structure appears on the web page as shown in Fig.8.
Fig.8. Screenshot of a benzene molfile as it appears on a web page using Chime. If you have the Chime plug-in, clicking on the image will open a new page with the fully interactive structure.
The resulting image can be manipulated in real time on the web page - it can be rotated around all 3 axes, the display style can be changed, it can be shown in stereo, and it can even be turned back into a 2D stick drawing, if required. Another excellent feature of Chime is the ability to run 'Chime scripts', which allow the author to alter the way different parts of the molecule are displayed. For example, here is the code to display the Chime structure of the chlorophyll pdb file shown in Fig.9.
<embed src="chlorophyll.pdb" width=220 height=220 startspin=true spiny=20 bgcolor="#AAFFFF" name="chlorophyll" csml="all cpk wireframe 50; atomno=01 cpk spacefill 200">
The new parameter 'csml' (Chime script mark-up language) is a string of other commands defining a set of instructions that Chime should perform when displaying the molecule. In this case, the first command states that to begin with, all the atoms in the molecule should be displayed as wireframe size 50, and be coloured according to the standard CPK convention [6]. The second command (which supercedes the previous one) says that atom number 1 (which is the Mg in the centre) should be displayed as a spacefill, sized 200. So this highlights the Mg at the centre of the molecule.
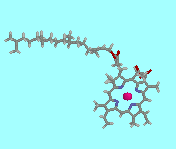
Fig.9. Screenshot of the Chime image of chlorophyll, displayed according to the Chimescript given in the main text. If you have the Chime plug-in, clicking on the image will open a new page with the fully interactive structure.
Longer, more complicated Chime scripts, can be saved as separate files (with extension csm or csml) which can then be read by the main program. An excellent example of the use of Chime scripts is to allow the reader to use 'push buttons' to highlight or alter certain parts of the molecule. Examples of the code for this can be seen below:
<embed src="chlorop2.csm" width=12 height=12 target="chlorophyll" button=push> Highlight the porphyrin ring</embed>
The chimescript called 'chlorop2.csm' (shown below) operates on the 'target' structure named 'chlorophyll' when the push button (size 12x12 pixels) is pressed by the reader. The Chime script first of all normalises the molecule display to wireframe size 50. The next line selects the Mg atom (atom number 1) and highlights it by making it larger - spacefill size 200). The subsequent lines select the individual atoms which make up the porphyrin ring, and colours each of them violet. Thus, when the script is activated by clicking on the button, the porphyrin ring stands out from the molecule, as shown in Fig.10.
|
all CPK wireframe 50 atomno=01 cpk spacefill 200 atomno=02 violet wireframe 50 atomno=03 violet wireframe 50 atomno=04 violet wireframe 50 atomno=05 violet wireframe 50 atomno=06 violet wireframe 50 atomno=07 violet wireframe 50 atomno=08 violet wireframe 50 atomno=09 violet wireframe 50 atomno=10 violet wireframe 50 atomno=17 violet wireframe 50 atomno=18 violet wireframe 50 atomno=19 violet wireframe 50 atomno=20 violet wireframe 50 atomno=21 violet wireframe 50 atomno=25 violet wireframe 50 atomno=26 violet wireframe 50 atomno=27 violet wireframe 50 atomno=28 violet wireframe 50 atomno=29 violet wireframe 50 atomno=33 violet wireframe 50 atomno=34 violet wireframe 50 atomno=35 violet wireframe 50 atomno=36 violet wireframe 50 atomno=37 violet wireframe 50 atomno=39 violet wireframe 50 atomno=41 violet wireframe 50 |
Fig.10. Example of a csml file, 'chlorop2.csm' above left. On the right is a gif screenshot of the interactive Chime image of chlorophyll, which includes push-buttons to highlight selected parts of the molecule. If you have the Chime plug-in, clicking on the image will open a new page with the fully interactive structure. The other csml files are: chlorop1.csm, chlorop2.csm, chlorop1.csm and chlorophyll.csm, which can be examined by clicking on the link and then viewing the page source in the browser.
Another browser plug-in which can read and display standard 3D structure files is ChemDraw3d Net. This operates in a very similar fashion to Chime, but is preferred by some authors who are used to the stand-alone ChemDraw package.
To avoid the need for a reader to download and install a separate plug-in package, an alternative method to display 3D structures on a web page is to use java applets. The most popular of these is called Chemsymphony, and it puts the onus of downloading the extra software upon the information provider, rather than the reader. The package is situated on the web server as a series of independent programs or 'beans', and the reader only requires a java-enabled browser to activate the packages. The java beans perform different processes, such as a 3D interactive display, measurement of atom-to-atom distances, displaying different molecules sequentially in the same window, etc. Again, the 3D coordinate data are stored in standard file formats (such as mol or pdb) and a series of applet parameters define how the image is to be displayed. The most basic applet is the 3D viewer called 'RenderBasic.class', and example HTML code to render a molfile on a web page is shown below. The parameter 'codebase' shows the path to the directory on the server which houses the Chemsymphony files. Other parameters are self-evident, and describe the type of display, and the size of atoms and bonds.
Fig.11. Example HTML code to render the Chemsymphony 3D display applet 'RenderBasic.class' (left), and a screenshot from the resulting 3D display for the aspartame molecule (right). If you have a java-capable browser, clicking on the image will open a new page with the fully interactive structure.
There are a number of stand-alone VRML viewers and browser plug-ins. The two most well known are Cosmoplayer and Cortona. These display the VRML (wrl) files in a fully interactive manner, and the user can rotate, walk or fly through the structure by clicking on buttons around the screen and/or dragging the mouse cursor. It is possible, using java or javascript controls, to open a VRML structure within an existing web page window. However, it is easier to open the VRML structure in a new window, since this is achieved by simply hyperlinking the wrl file to selected words or images within the web page. An example of the screenshot of the resulting VRML file for aspartame (as displayed in CosmoPlayer) is shown in Fig.12, and if you have a VRML plug-in, clicking on the image will open the apartame wrl file in a new window in your browser.
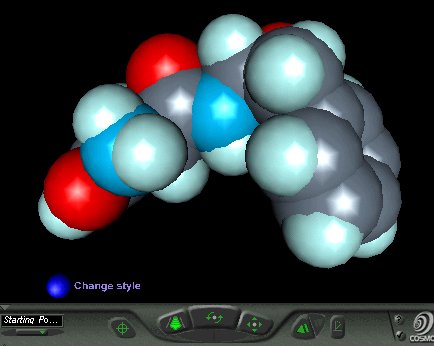
Fig.12. Screenshot of the aspartame VRML file as displayed in spacefill style in CosmoPlayer. If you have a VRML plug-in, clicking on the image will open the fully interactive file in a new window.