

For comments or information, please contact R. Frey at gfrey@wuchem.wustl.edu.
NOTE: Click on text that is highlighted in blue for more information on that topic.
Iron, an essential element in living organisms, is commonly used in the Fe(II) oxidation state. But in our oxidizing atmosphere, Fe(III) is the more prevalent oxidation state. At the physiological pH of 7, the Fe(III) ion concentration in aqueous solution is minimal. However, most organisms maintain an intracellular concentration of Fe(III) several orders of magnitude higher than simple aqueous solutions permit. This discrepancy in concentration demonstrates the striking ability of biochemical systems to concentrate and store iron. Conversely, iron can be very toxic, so the ability to store and release iron in a controlled fashion is essential. Cells have solved this problem of iron storage by developing ferritins, a family of iron-storage proteins that sequester iron inside a protein coat as a hydrous ferric oxide-phosphate mineral similar in structure to the mineral ferrihydrite.
This graphical tutorial is used in conjunction with a general-chemistry laboratory experiment which explores the in vitro iron-release process. Pdb files of the compounds used in the experiment are available at the bottom of this tutorial for downloading and interactively viewing using an appropriate molecular-viewing package such as Rasmol. Below are four (4) points of emphasis within the tutorial:
Ferritin is a spherical shell that consists of 24 subunits (or peptide chains) folded into ellipsoids, as shown in Figure 1. Each subunit is an individual molecule that joins to its neighboring subunits through noncovalent interactions and have a combined molecular weight of 474 000. These subunits pack to form a hollow sphere approximately 80 Angstroms in diameter with walls that are approximately 10 Angstroms thick. Among the important structural features of ferritin is the presence of two types of channels that occur in the protein wall at the intersection of the subunits.
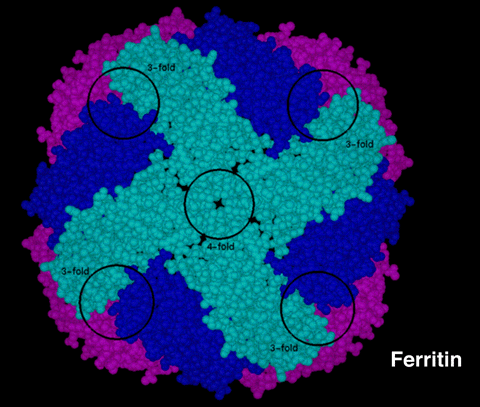 |
Figure 1:This is a molecular model of Ferritin with the subunits displayed in the CPK representation. (All of the 24 subunits are identical, but they have been color coded to help illustrate the structure. Light blue subunits are closest to you, magenta subunits are farthest away, and dark-blue subunits are in-between.) Coordinates for the model were determined from X-ray crystallographic data. The four (4) subunits colored in light blue form the walls of a 4-fold channel. The 3-fold channels occur at the intersection of the light blue, dark blue, and magenta-colored subunits. The location of 3-fold channels are indicated on the figure, but the channels themselves are obscured from this viewing angle. |
Questions concerning the background of ferritin
In vitro studies on the removal of iron from ferritin are of interest because of ferritin's possible use in the treatment of iron overload and in increasing our understanding of iron metabolism. Below are two schemes that outline the in vitro iron-removal mechanism, as performed in the general-chemistry experiment.
NOTE: These schemes are not drawn to scale.
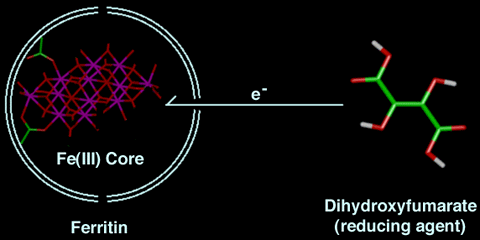 |
Scheme 1:This is a two-dimensional representation of the first step of the reaction. The iron-mineral core is attached to the ferritin shell. The protein wall is 10 Angstroms thick and is denoted by the double white line. The diameter of the inside of the protein shell is 80 Angstroms and the channels are represented by breaks in the protein shell. DHF may enter through a 4-fold channel (hydrophobic) or may transfer an electron to the mineral core via another mechanism. (Remember the compounds in this scheme are not drawn to scale.) Note: The carbon atoms are green, the oxygen atoms are red, the hydrogens are white, and the iron atoms are magenta in the stick representations. |
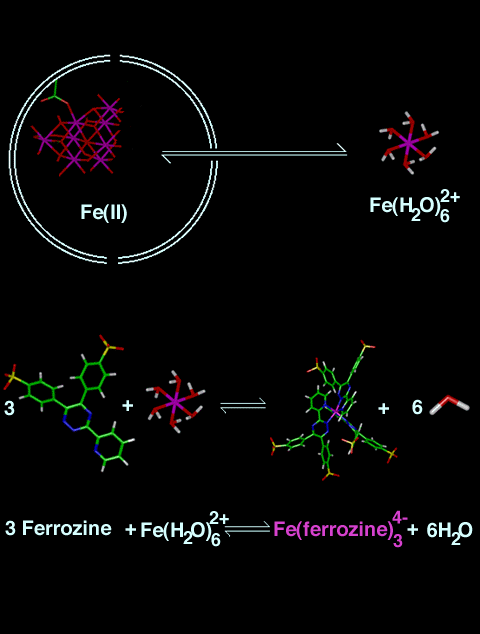 |
Scheme 2:This is a two-dimensional representation of the remaining steps in the reaction. After Fe(III) is reduced to Fe(II), Fe(II) leaves the protein via a 3-fold channel and reacts with three (3) dianionic ferrozine ligands outside the protein shell. The reaction of Fe(II) with ferrozine2- results in formation of the Fe(ferrozine)34- compound. (Remember the compounds in this scheme are not drawn to scale.) Note: The carbon atoms are green, the hydrogens are white, the iron atoms are magenta, the nitrogen atoms are blue, the oxygen atoms are red, and the sulfur atoms are yellow in the stick representations. |
Figure 2 contains all of the main species involved in the in vitro iron-removal process described in this tutorial. This figure allows you to compare the relative sizes of the different systems involved. Clicking on each of the species in the picture also links you to a more detailed picture of the respective compounds. When you are looking at the species in detail, you should be predicting what the geometric structure would look like based on your intuition and how that structure affects the role that each species plays in the iron-removal mechanism?
![[Imagemap]](glossary_net.gif)
|
Figure 2:This is a molecular model representation depicting the relative sizes of the main species involved in the reaction. To see a more detailed picture of each species, please click on each of the species in the picture. (Note: There is not a more detailed picture of the protein.) Starting at the top of the figure on the right-hand side are Dihydroxyfumarate, Fe(II) hydrated complex, Ferrozine 2- ligand, and Fe(ferrozine)34-. The iron-mineral core is shown inside the protein shell. |
To view the molecules interactively, please use RASMOL. To download the pdb files for viewing and rotating the compounds used in this experiment, please click on the appropriate compound name below. (Note: If you want to view multiple compounds simultaneously, please download each pdb file and save on your computer. Then open RASMOL outside of Netscape.)
To download the pdb files corresponding to some of the components of the protein, please click on the appropriate molecule name below:
Harrison, P.M.; Andrews, S.C.; Artymiuk, P.J.; Ford, G.C.; Guest, J.R.; Hirzmann, J.; Lawson, D.M.; Livingstone, J.C.; Smith, J.M.A.; Treffry, A.; Yewdall, S.J. in Iron Transport and Storage; Ponka, P.; Schulman, H.M.; Woodworth, R.C., Eds.; CRC: Boca Raton, FL, 1990; pp 81-101.
Theil, E.C. Annu. Rev. Biochem.1987, 56, 289-316.
Lawson, D.M.; Artymiuk, P.J.; Yewdall, S.J.; Smith, J.M.A.; Livingstone, J.C.; Treffry, A.; Luzzago, A.; Levi, S.; Arosio, P.; Cesareni, G.; Thomas, C.D.; Shaw, W.V.; Harrison, P.M. Nature 1991, 349, 541-544.
Acknowledgements: The development of this experiment was supported by the National Science Foundation Division of Undergraduate Education Grant# NSF/DUE-9455918 to the ChemLinks program and by a grant from the Howard Hughes Medical Institute through the Undergraduate Biological Sciences Education program, Grant HHMI# 71195-502005 to Washington University. The molecular-modeling calculations were performed using the Washington University Department of Chemistry Computer Facility, which was funded in part by Washington University's Parents' Fund.
Copyright 1995, Washington University, All Rights Reserved.